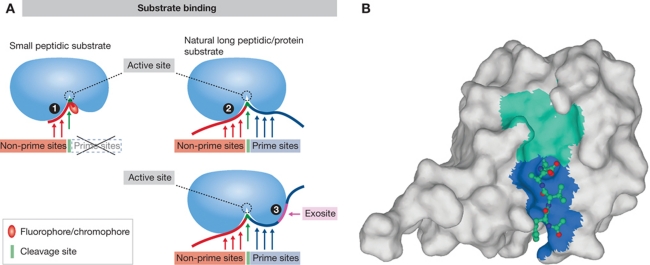
Figure 1.
Binding of a substrate to a protease. (A) Schematic representation of substrate binding. The basic differences between binding of a small peptidic or peptidomimetic substrate and a larger peptide or protein substrate are shown. Small substrate binds tightly to the non-prime binding sites whereas the usually bulky fluorophore/chromophore (leaving group) binds to the S1′ site while the other prime sites remain empty (1). Larger peptidic substrates bind to both prime site and non-prime site binding sites, while the interaction with some individual binding sites can be looser (2). Exosites on protease surface serve as additional binding elements for large substrates to strengthen the interaction with the protease and allow recognition (3). They can be also used for discrimination between the different substrates. (B) Small substrate-mimicking inhibitors that helped tremendously in elucidating the substrate binding mechanism(s) often bind the same way as the small substrates (some more differences with metalloproteases), except that in covalent inhibitors the leaving group is replaced by a warhead (see A for a schematic representation). Crystal structure of caspase-1 in complex with the inhibitor YVAD-CHO (Wilson et al, 1994; 1ICE) revealed the insight into the mechanism of substrate binding to the active site of caspases as an example of such small molecule (substrate or inhibitor) binding. The surface of caspase 1 is shown in light grey. In the active site cleft, the non-primed and primed parts are shown as cyan and blue surface. The structure of the inhibitor (shown as a ball-and-stick model) has revealed the S1, S2, and S3 substrate binding sites. The non-hydrogen atoms C, N, and O are shown in blue, dark blue, and red, respectively. Figure was prepared with MAIN (Turk, 1992) and rendered with POV-Ray.