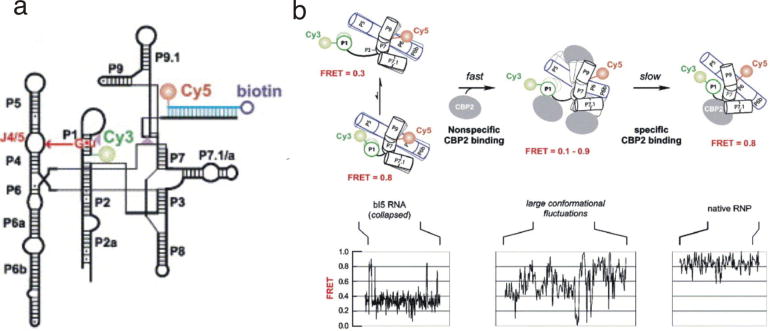
Fig. 6.
Single molecule study of the bI5 group I intron and its folding dynamics in the presence of CBP2. (a) The secondary structure of the bI5 group I intron showing positions of Cy3, Cy5, and biotin. (b) The proposed two-step model for the formation of bI5-CBP2 complex (upper panel) and the respective single molecule FRET trajectories (lower panel). In the absence of the protein (left), the RNA collapses to a 0.3 FRET state and makes transient excursions to a 0.8 FRET state with native-like features. In the presence of CPB2, a rapid and non-specific binding occurs first, which cause a large conformational fluctuation as shown by the single molecule FRET trajectories (middle). Finally, the slow and specific binding of CBP2 protein to RNA forms a stable (0.8 FRET state) and catalytically active CBP2-bI5 complex (right). This figure was reprinted from (47) with permission from Elsevier.