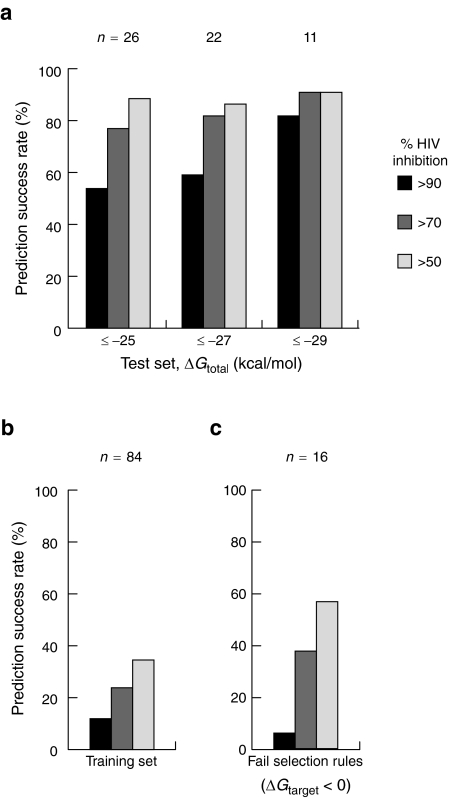
Figure 6.
Prediction success rates of designed shRNAs. (a) Inhibition of HIV-1 production by shRNAs in the test set. Target sequences were chosen to have accessible 13-nucleotide windows (ΔGtarget ≥0 kcal/mol) and strong overall binding energy ΔGtotal. Prediction success rates are shown for ΔGtotal criteria of -25, -27, and -29 kcal/mol; n, the numbers of shRNAs meeting a given threshold. (b) Inhibition of HIV-1 production by shRNAs in the 84-member training set. (c) Prediction success rates for 16 shRNAs that failed the target accessibility criterion. ΔGtarget <0 corresponds to RNA target sites with some pre-existing structure.