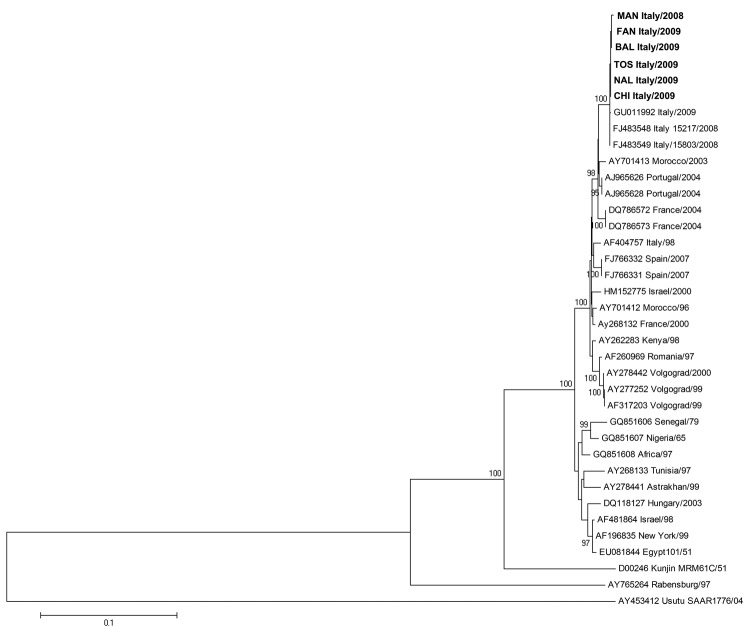
Figure 1.
Phylogenetic trees of West Nile virus strains isolated during outbreaks in Italy, 2008–2009, based on nucleotide sequences of the complete envelope gene. Phylogenetic tree and distance matrices were constructed by using nucleotide alignment, the Kimura 2-parameter algorithm, and the neighbor-joining method implemented in MEGA version 4.1 (www.megasoftware.net/mega4/mega41.html). The tree was rooted by using Usutu virus as the outgroup virus. The robustness of branching patterns was tested by 1,000 bootstrap pseudoreplications. The percentage of successful bootstrap replicates is indicated at nodes, showing only values >80. The scale bar indicates nucleotide substitutions per site. Strains sequenced in this study are TOS-09, FAN-09, BAL-09, CHI-09, NAL-09, and MAN-08. Envelope nucleotide sequence GenBank accession nos: NAL-09, HM991272; TOS-09, HM991273; BAL-09, HM991274; CHI-09, HM991275; MAN-09, HM991276; FAN-09, HM991277. Boldface indicates the 6 newly sequenced strains.