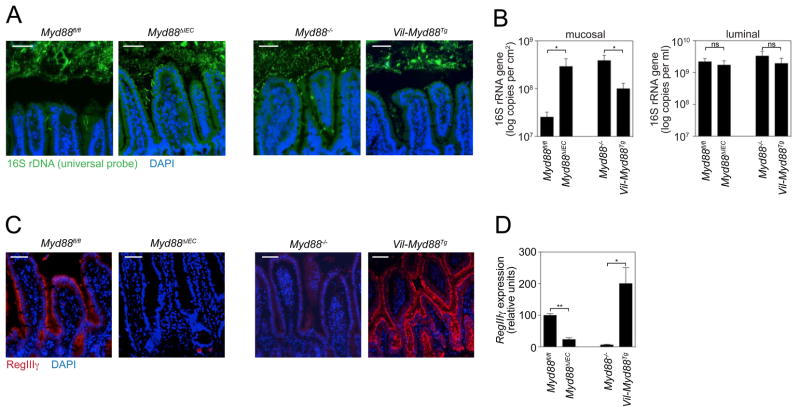
Fig. 2.
Epithelial cell MyD88 is necessary and sufficient to limit bacterial association with the small intestinal surface. Myd88 ΔIEC and Vil-Myd88Tg mice were each analyzed alongside co-housed littermates (Myd88fl/fl and Myd88−/−, respectively). (A) FISH analysis of microbiota localization in terminal ileum using a universal bacterial 16S rRNA gene probe. Images are representative of >10 littermate groups. (B) Mucosa-associated and luminal bacteria were quantified by Q-PCR determination of 16S rRNA gene copy number in the terminal ileum. N=5 mice/genotype; data are from three littermate groups. (C) RegIIIγ was detected in distal small intestine with anti-RegIIIγ antiserum (13) (red). Nuclei were counterstained with DAPI (blue). Images are representative of three littermate groups. (D) Q-PCR quantification of RegIIIγ transcripts in terminal ileum (N=5 mice/genotype from three littermate groups). Scale bars=50 μm; *, p<0.05; **, p<0.01; Error bars, ±SEM; ns, not significant.