Figure 1.
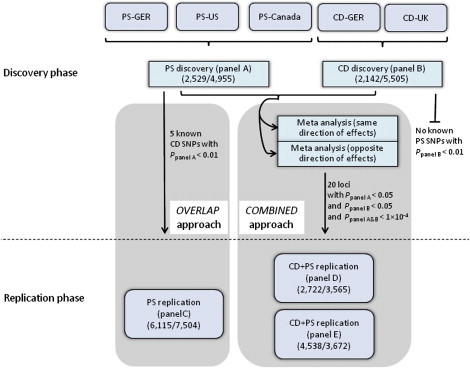
Study Design for the Combined Analysis of CD and PS
For discovery, we conducted a PS and a CD GWAS meta-analysis (panel A and B in Table S1), respectively, and employed two strategies (OVERLAP and COMBINED) to systematically search for shared risk loci. In a first approach (OVERLAP), we tested established non-HLA PS risk SNPs for potential association (p < 0.01) with CD and vice versa. In a second approach (COMBINED), we selected SNPs from 20 loci for being nominally associated in each of the single-disease meta-analyses (ppanel A < 0.05, ppanel B < 0.05) and for being significantly associated in the combined-phenotype association analysis at the 10−4 level (ppanel A&B < 10−4). For replication, follow-up SNP genotyping was performed for CD and PS in independent replication panels (panels C–E in Table S1). The following abbreviations are used: PS-GER, German PS GWAS; PS-US, United States PS GWAS; PS-Canada, Canadian PS GWAS; CD-GER, German CD GWAS; and CD-UK, United Kingdom CD GWAS. For each panel, numbers of cases/controls are displayed in parentheses.
