Figure 3.
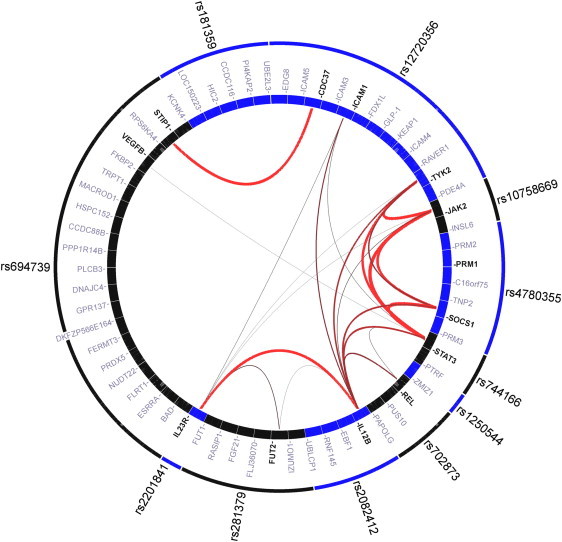
Gene Relationships across the 11 Shared Risk Loci of CD and PS Identified by GRAIL Analysis
GRAIL46 is a statistical text-mining approach to quantify the degree of relatedness among genes in genomic disease regions. It estimates the statistical significance of the number of observed relationships with a null model in which relationships between the genes occur by random chance. A significance score ptext, which is adjusted for multiple hypothesis testing, represents the output GRAIL score. ptext values approximately estimate type-I error rates. Outer circle: lead SNPs from shared risk loci of both diseases; each box represents a SNP. Inner circle: genes of the genomic regions around lead SNPs that were identified based on LD properties; each box represents a gene; genes that were scored at ptext < 0.05 are significantly linked to genes in the other disease regions and are indicated in bold type. Lines: the lines between genes represent significant connections, with the thickness and redness of the lines being inversely proportional to the probability that a text-based connection would be seen by chance.
