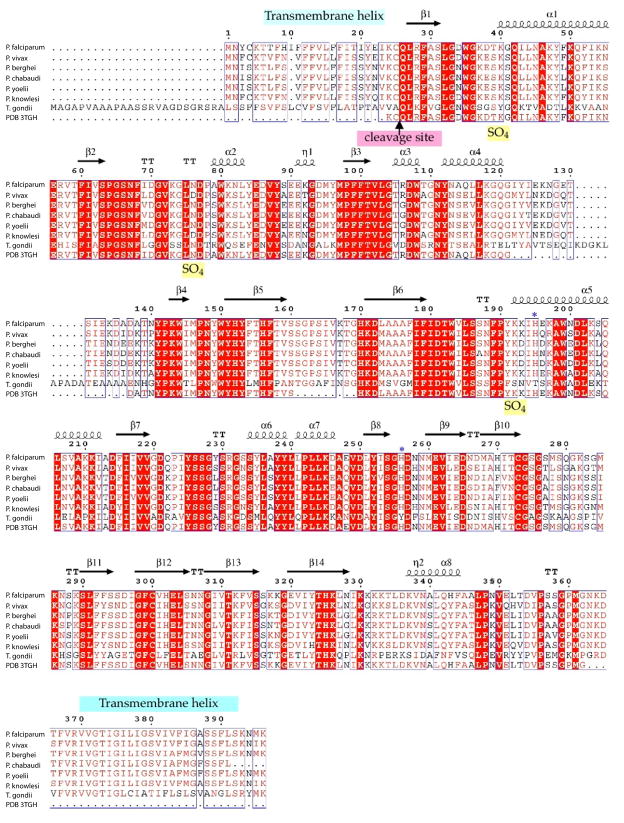
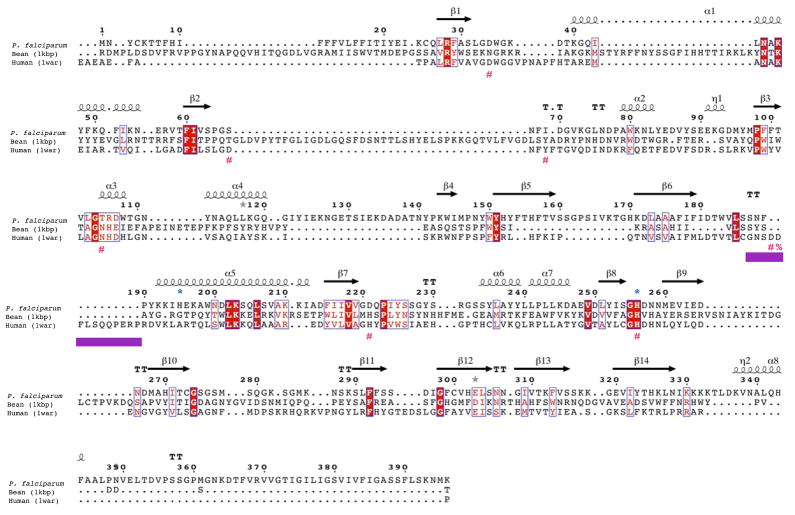
Figure 2. GAP50 sequence alignments.
A) GAP50 sequence alignment of various Plasmodium species and Toxoplasma gondii. (sequence information was obtained through BLAST searches in EuPathDBhttp://eupathdb.org). Predicted transmembrane helices are indicated in light blue bars. The blue *symbols represent residues involved in coordination of the cobalt ions, the Toxoplasma (Daher and Soldati-Favre, 2009) and Plasmodium signal sequence cleavage site (Müller et al., 2010)(Yeoman et al., 2011) is indicated by an arrow.
B) Structure based sequence alignment of Plasmodium falciparum GAP50 Human PAP and kidney bean PAP. The secondary structure elements of the Plasmodium GAP50 structure is shown above the sequence. The repression loop of hPAP is highlighted with a purple box and is inserted between Ser187 and Asn188 of the Plasmodium sequence. The key residue for inhibition of the human enzyme is marked with a red %. The blue *symbols represent residues involved in coordination of the cobalt ions in PfGAP50, red #symbols represent active site residues coordinating the iron atoms in hPAP.