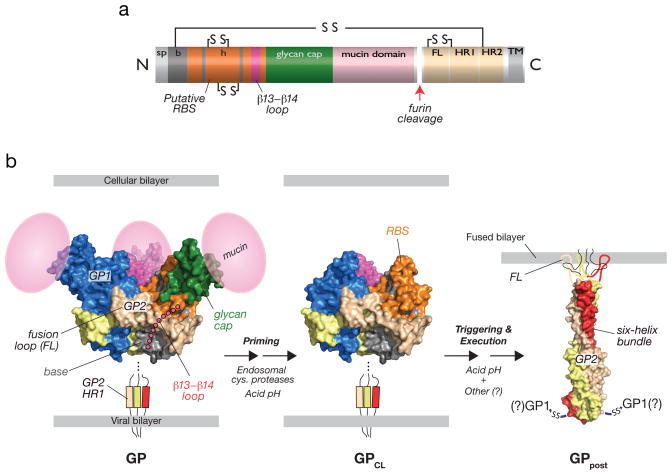
Fig. 1. Structural organization and features of the filovirus glycoprotein, GP, and proposed structural transitions during entry.
(a) Linear diagram of GP indicating sequence features. The cellular Golgi endopeptidase furin cleaves the precursor subunit GP0 to GP1 and GP2. sp, signal peptide. b, GP1 ‘base’ subdomain. h, GP1 ‘head’ subdomain. RBS, putative receptor-binding sequence. FL, GP2 fusion loop. HR1, GP2 N–terminal heptad repeat. HR2, GP2 C–terminal heptad repeat. TM, GP2 transmembrane domain. Lines and ‘SS’ indicate intra- and intersubunit disulfide bonds. (b) Structures of an ectodomain of the EBOV GP pre-fusion trimer (PDB id: 3CSY [9]), GP2 in its putative post-fusion conformation (PDB id: 1EBO [7]), and a structural model of a proteolytically-primed GP intermediate (GPCL). The β13-β14 loop connecting the GP1 base and head subdomains, and the GP1 mucin domain were not visualized in the pre-fusion crystal structures (3CSY and 3S88 [10]) and are shown as cartoons. The GP2 fusion loop was not visualized in the post-fusion crystal structues (1EBO and 2EBO [8]) and is shown as a cartoon. During entry, endosomal cysteine proteases cleave and remove the glycan cap and mucin subdomains of GP1, generating GPCL, in which the putative rbs is fully exposed. In response to an unknown host trigger, GP2 is proposed to undergo release from its pre-fusion conformation and to rearrange to form the six-helix bundle. Formation of this structure is thought to drive fusion of the viral and cellular lipid bilayers. The disposition of GP1 and the intersubunit disulfide bond during membrane fusion and in this post-fusion GP conformer (GPpost) is unknown. Acid pH is required for endosomal cysteine protease activity and is hypothesized to play multiple roles in fusion (see text for details).