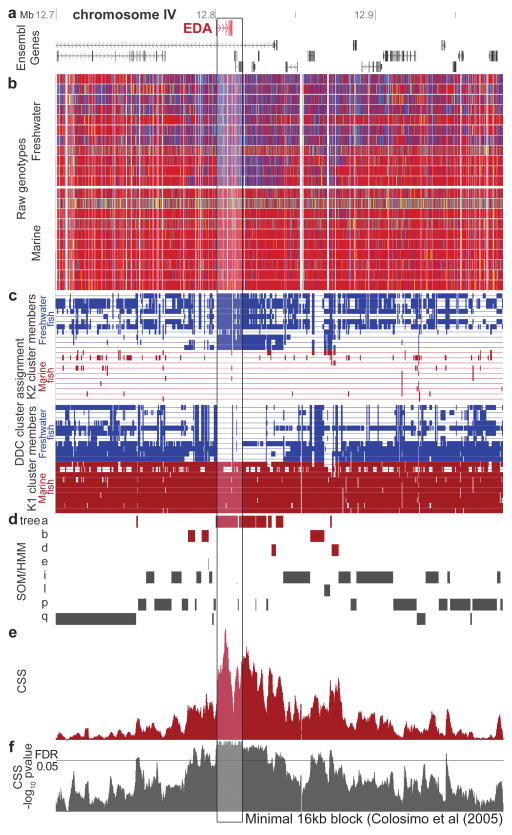
Figure 2. Parallel divergence signals at known armour plates locus.
a. Ensembl gene models around EDA. b. Visual genotypes for sequenced fish [homozygous sites for most frequent allele in marine fish (red); homozygous for alternate allele (blue); heterozygous (yellow), or nonvariable/missing/repeat-masked data (white)]. c. DDC cluster assignments for marine (red) and freshwater populations (blue). Most fish are assigned to cluster k1, except in boxed region, where freshwater fish are assigned to a distinct cluster (k2). d. SOM/HMM analysis supports patterns of divergence with a marine-freshwater-like tree topology in the centre, but not edges, of the window (trees a-d). Similar support is shown by CSS analysis (e) and its associated P-value (f). The combined analyses define a consensus 16 kb region shared in freshwater fish (vertical shaded box), matching the minimal haplotype known to control repeated low armour evolution in sticklebacks11.