Abstract
The p53 pathway displays a large degree of redundancy in the expression of a number of pro-apoptotic mechanisms following DNA damage that, among others, involves increased expression of several pro-apoptotic genes through transactivation. Spatial and temporal cellular contexts contribute to the complexity of the regulation of apoptosis, hence different genes may show a cell- and tissue-dependent specificity with regard to the regulation of cell death and act in concert or show redundancy with one and another. We used siRNA technology to assess the effect of multiple ablations of documented pro-apoptotic p53 target genes (PPG) in the colorectal cancer cell line HCT116 and generated mice deficient in both of the extrinsic and intrinsic PPGs genes Dr5 and Puma following treatment with chemotherapeutics and ionizing radiation. DR5, Fas, Bax, Bad, Puma and Bnip3L were induced by 5-FU and adriamycin (ADR) in HCT116 cells in a p53-dependent manner. The resulting caspase 3/7 activity in HCT116 cells following treatment were suppressed by ablated expression of the PPGs in the extrinsic as well as the intrinsic pathway. To our surprise, knocking-down any of the PPGs concomitantly with DR5 did not further inhibit caspase 3/7 activity whereas inhibiting DR5-expression in HCT116Bax knockdown (kd) and HCT116Fas kd did, suggesting that these genes act downstream or in synergy with DR5. This was supported by our in vivo observations, since Puma and Dr5 were equally efficient in protecting cells of the spleen from sub-lethal radiation-induced apoptosis but less effective compared with irradiated p53−/− mice. To our surprise, Dr5−/−; Puma−/− mice did not show additive protection from radiation-induced apoptosis in any of the investigated organs. Our data indicates that the intrinsic pathway may rely on extrinsic signals to promote cell death in a cell- and tissue-dependent manner following DNA damage. Furthermore, p53 must rely on mechanisms independent of DR5 and PUMA to initiate apoptosis following γ-radiation in the spleen and thymus in vivo.
Key words: p53, KILLER/DR5, PUMA, apoptosis, DNA damage
Introduction
The p53 protein plays a critical role in inhibiting carcinogenesis, as half of all human tumors carry inactivating mutations in the p53 gene, and the other half may have dysfunctional p53 as a consequence of alterations in post-translational events or inhibition of upstream activators.1 Under normal physiological conditions, the level of p53 are kept low and functionally switched off, whereas stressors, such as DNA damage, oncogenic signaling, hypoxia or nucleotide depletion, result in activation of p53.2 Activated p53 accumulates as a tetramer in the cell nucleus and act as a transcription factor for genes involved in cell cycle arrest and programmed cell death (apoptosis).3 In the case of cell cycle arrest, p21 appears sufficient to block cell cycle progression in G1 phase until repair of damaged DNA has occurred or the cellular stress has been resolved.4 However, the p53-dependent apoptotic response is more complex and involves transcriptional activation of multiple pro-apoptotic target genes that display tissue, cell and context-dependent specificity in their ability to execute apoptosis. p53 transactivates key molecules in several apoptosis pathways, including Fas, DR4, KILLER/DR5, DcR1, DcR2, Bid, Puma, Noxa, Bad, Bak, Bax, p53AIP1, caspase 6, Apaf1 and Bnip3L (also referred to as Nix)5,6 and may also transrepress anti-apoptotic genes such as Bcl-2 and Survivin.7,8
The use of mice lacking functional p53 has made the existence of a DNA damage-induced type of apoptosis evident, which presents itself only in the presence of functional p53. p53-null mice are resistant to apoptosis induced by γ-radiation in the developing nervous system,9 spleen, thymus10 and the small intestine.10–12 Additionally, p53-deficient mice are resistant to apoptosis triggered by 5-fluorouracil (5FU) in the small intestine,13 arabinofuranosylcytosine in sympathetic neurons14 and by adriamycin in the thymus, spleen and small intestine.10 However, loss of either Bax or Bak alone is not sufficient to confer resistance to apoptotic stimuli, whereas cells lacking both Bax and Bak become resistant to many types of cellular stress.15 In contrast, Puma-null thymocytes, developing neurons and hematopoietic cells do not trigger apoptosis following DNA damage.16,17 HCT116 Puma−/− cells are resistant to apoptosis induced by p53 overexpression.18 P53-triggered death receptor induction may also play a role in DNA damage-triggered apoptosis, since Dr5-deficient mice show reduced amounts of apoptotic cells in their thymus, spleen and white matter of the brain following ionizing radiation.19 These studies have established the importance of the p53-dependent pro-apoptotic target genes in the execution of apoptosis. However, it is largely unclear to what extent PPGs show redundancy and interdependency, perhaps due to the tedious nature of generating mice deficient in multiple genes.
Here, we address the possibility of synergy between proapoptotic proteins in the p53 pathway by employing siRNA to target the expression of several PPGs and subsequently screen for caspase 3/7 activity following chemotherapy in HCT116 colorectal cancer cells. We also generated mice lacking both Dr5 and Puma and subjected these mice to exposure to sub-lethal doses of ionizing radiation in order to evaluate the concomitant role of these two pro-apopototic molecules in p53-dependent apoptosis in normal radio-sensitive tissues in vivo. Our results indicate a previously unknown interdependency between key pro-apoptotic genes involved in the intrinsic (mitochondrial) and extrinsic pathway for the execution of DNA damage-induced apoptosis.
Results
5FU triggers p53-dependent cell death in HCT116 and PA-1 cells.
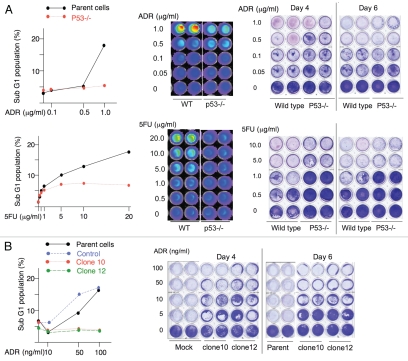
To study p53-dependent apoptosis in human cancer cells, we screened a number of cell lines [PA-1, U2OS (osteosarcoma cells), H460 (lung cancer cells), HCT116] expressing wild-type p53 with a number of chemotherapeutics (5FU, Adriamycin [ADR], cyclophosphamide, gemcitabine) known to cause DNA damage for a combination that triggered p53-dependent cell death. In order to verify the dependency for p53 in the execution of apoptosis, we silenced p53 in PA-1, U2OS and H460 cells using micro RNA against p53 (Sup. Fig. 1B and data not shown). We used HCT116-p53−/− cells for their counterpart, as we could not obtain efficient p53 knockdown in the HCT116 cell line (Sup. Fig. 1A). We were able to confirm that both 5FU and ADR induced p53-dependent cell death in HCT116 cells and adriamycin in PA-1 cells by sub-G1 analysis and long-term cell culture survival following exposure (Fig. 1). The other combinations of cells and chemotherapeutics failed to induce apoptosis associated with reduced long-term cell culture survival or did so independently of p53. Based on these results, we used HCT116 and PA-1 cells to study p53-dependent cell death.
Figure 1.
Induction of p53-dependent cell death in HCT116 and PA-1 cells. Cells were treated with different concentrations of adriamycin or 5FU for 16 h and subjected to sub-G1 analysis (left) and long-term cell survival assay as described in the Materials and Methods section (right). HCT116 and HCT116-p53−/− cells (A) and PA1 cells and the clones of p53-knockdown cells (B).
DR5, Fas, Bax, Bad, Puma and Bnip3L were induced by 5FU and adriamycin in HCT116 cells in a p53-dependent manner.
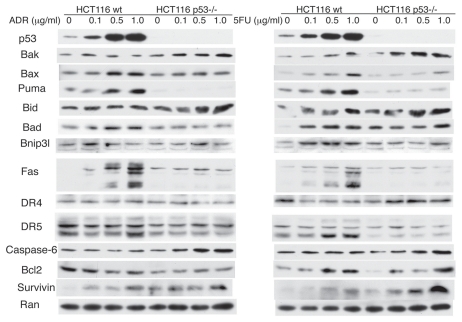
We next studied the expression of PPGs or anti-apoptotic genes that are reported to be repressed by p53 under the previously established conditions triggering p53-dependent cell death. As shown in Figure 2, DR5, Fas, Bax, Bad, Puma and Bnip3L were induced by 5FU and ADR in HCT116 cells in a p53-dependent manner. In contrast, the expression levels of Noxa and Apaf1 were very low in this particular cell line, and there were no appreciable change after treatment with ADR or 5FU. As we lacked a good antibody for p53AIP1, we performed semi-quantitative RT-PCR to analyze the expression of the gene following treatment with chemotherapeutics. However, we were unable to find conditions where this gene could be upregulated by 5FU or ADR in a p53-dpendent fashion (Sup. Fig. 2). Based on these results, we decided to target DR5, Fas, Bax, Bad, Puma and Bnip3l for multiple-gene silencing, utilizing siRNA to study the relationship among the genes and their contribution to the p53 apoptotic response.
Figure 2.
Induction of p53 target genes by ADR and 5FU HCT116 and HCT116 p53−/− cells were treated with ADR or 5FU for 16 h and the cell lysates were subjected to protein gel blot analysis for known p53 target genes.
Fas and DR5 triggers p53-dependent apoptosis following 5FU treatment in colorectal cancer cells in vitro.
We inhibited expression of the p53-inducible pro-apoptotic genes alone and in combinations and assessed caspase 3/7 activity using the caspase Glo assay following treatment with chemotherapy. We treated the cells with 20 µg/ml of 5FU, since these conditions showed p53-dependent caspase 3/7 activation in the preliminary experiments. The effect of gene silencing was confirmed by protein gel blot (Sup. Fig. 3).
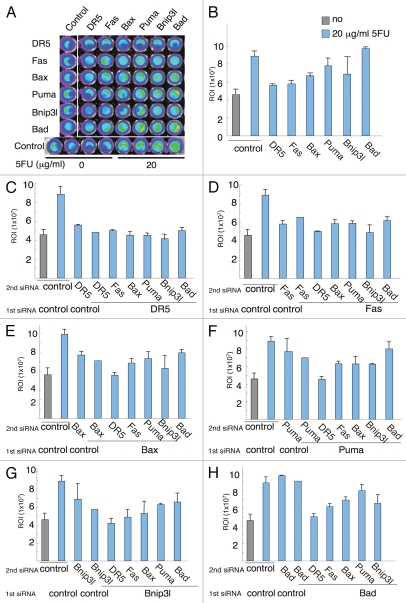
The result of single gene silencing suggested that, not only did genes involved in the initiation of apoptosis at the level of mitochondria participate in the activation of caspase 3/7 following treatment with 5FU, but so did the members of the TNF death receptor family (DR5 and Fas, Fig. 3B). Silencing of DR5 displayed the greatest inhibition (80%) of caspase 3/7 activity following 5FU of all the genes tested (Fig. 3B). This was somewhat surprising but consistent with what has been reported earlier with respect to death receptor signaling following DNA damage.19–23
Figure 3.
The effects of siRNA silencing of pro-apoptotic p53-dependent target genes (PPG) on caspase 3/7 activity in HCT116 cells. The expression of the PPGs was blocked by siRNA in the colorectal cancer cell line HCT116 indicated and treated with 20 µg/ml 5FU for 16 h (A). Caspase 3/7 activity was measured using the IVIS system. The signals from the plate were measured and are shown as bar graphs. Results are the summary of two independent experiments (n = 4). (B) Single gene knockdown. Combination gene silencing of DR5 (C), Fas (D), Bax (E), Puma (F), Bnip3L (G), Bad (H) and the other p53 target genes.
We further analyzed the results of concomitant gene silencing in order to address the relative impact and potential synergy between genes that trigger p53-dependent apoptosis. We found that only Bax, Puma and Bnip3L displayed additive inhibition effects on caspase 3/7 cleavage to that of DR5 knockdown (Fig. 3C). The combination reduced the level of caspase 3/7 activity to basal levels, suggesting that these genes participate in distinct pathways that trigger p53-dependent apoptosis in HCT116 cells following 5FU. In contrast, when silencing Fas in combination with the BH3-only pro-apoptotic proteins (Fig. 3D), concomitant inhibition of Bnip3L was most effective in inhibiting caspase 3/7 activity. Silencing both DR5 and Fas also caused an additive inhibition of caspase 3/7 activation. Interestingly, ablating Fas expression on top of ablated DR5 expression did not cause any additive effect (Fig. 3C). This indicates that Fas-dependent caspase 3/7 cleavage may show unilateral dependency on the DR5-TRAIL-system. Indeed, it has been shown that Fas-induced liver damage is accelerated by the presence of TRAIL in mice in vivo,24 supporting a role for TRAIL in Fas-induced apoptosis. Furthermore, Fas-induced activation of caspase 3/7 showed a differential dependency on the BH3-only proteins compared with DR5, since no additive effect on caspase 3/7 cleavage following siRNA-inhibition of either Puma or Bax was detected after 5FU exposure (Fig. 3D).
Silencing of Bax and DR5 decreased the caspase activity to basal levels comparable to that of untreated cells (Fig. 3E). A similar synergy was also seen for DR5 and Puma (Fig. 3F) and DR5 and Bnip3L (Fig. 3G). When the effect of silencing combinations for Bax, Puma and Bnip3L were compared (Fig. 3E–G), there were no significant additive effects among these genes, suggesting they act on a redundant signal transduction pathway that triggers p53-dependent apoptosis in HCT116 colorectal cancer cells following 5FU, in concert with previously published data. We were unable to detect any effect of Bad silencing on caspase 3/7 cleavage following treatment with 5FU throughout the experiment (Fig. 3H).
Loss of Dr5 and Puma do not synergistically protect from cell death following ionizing irradiation in vivo.
In order to investigate synergy between the extrinsic and intrinsic (mitochondrial) cell death pathways in vivo, we generated mice lacking both Dr5 and Puma. Previous studies demonstrated that p53−/−, Dr5−/− and Puma−/− mice are protected from apoptosis following ionizing radiation.16,19,25 Neither Dr5−/− nor Puma−/− mice display any severe phenotype early in life and neither did mice lacking both Dr5 and Puma (i.e., Dr5−/−; Puma−/− mice) (data not shown). However, p53−/− mice developed malignancies at a rate and phenotype consistent with what has been previously described in reference 26. Thus, knocking out both Puma and Dr5 did not recapitulate the tumor susceptibility phenotype of p53-null animals even though they presumably would act through non-redundant, pro-apoptotic signal transduction pathways.
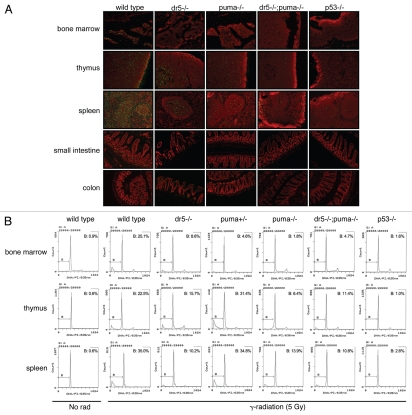
The p53-dependent pro-apoptotic response involves increased transcription of Dr527 and Puma,18 suggesting that apoptosis could be triggered following DNA damage by endogenous TRAIL.28 In order to investigate how the downstream DNA damage response was affected by loss of Dr5 and Puma, we subjected wild-type, Dr5−/−, Puma+/−, Puma−/− and Dr5−/−; Puma−/− and p53−/− female mice (4–6 weeks) to 5 Gy of whole-body irradiation. The number of apoptotic cells (as measured by TUNEL-staining and sub-G1 analysis) was markedly increased following irradiation in all tissues investigated relative to control (data not shown and Fig. 4B). Loss of p53 led to a substantial reduction in TUNEL-staining and sub-G1 fraction in both the lymphoid/hematopoietic organs as well as the GI-tract following irradiation (Fig. 4). This suggests that cell death following irradiation is highly, if not exclusively, dependent on p53 in the bone marrow, spleen and thymus at this time-point. Loss of Dr5 led to a reduction of TUNEL-positive cells/sub-G1 fraction in the bone marrow, thymus and a substantial reduction of TUNEL-positive cells in the white pulp of the spleen (Fig. 4), whereas only a very modest reduction in TUNEL-positive cells in the crypts in the small intestine and colon Dr5−/− animals were found. However, despite that DR5 mediates radiation-induced apoptosis in the hematopoietic and lymphoid organs in vivo, it is not the only p53-dependent mediator of cell death following irradiation.
Figure 4.
p53-, Puma- and Dr5-dependent apoptosis in radiosensitive organs in vivo. Mice 4–6 weeks of age were subject to whole-body irradiation (5 Gy) and sacrificed 8 h later. (A) Apoptosis was detected by TUNEL staining (FITC/green) counter stain by propidium iodide. (B) Apoptosis (sub-G1 population) was quantitated ex vivo by flow cytometry and sub-G1 analysis. Representative photographs and flow cytometry data from two independent experiments are shown.
Interestingly, loss of Puma in the bone marrow led to an equivalent reduction of cell death following irradiation compared with loss of p53 in the organ (Fig. 4B). This suggests that PUMA is an essential p53-dependent mediator of cell death in the bone marrow. Indeed this is also in agreement with previous findings that document increased survival of Puma-deficient animals to lethal doses of ionizing irradiation that trigger severe myelosuppression.16,29 Furthermore, a pronounced, organ-specific gene dose dependency was also observed for the puma-gene. One allele was sufficient to significantly quench apoptosis in the bone marrow (Fig. 4B), but this puma genotype offered no protection for radiation-induced apoptosis in the spleen and thymus. This indicates an organ-specific mechanism for PUMA in the triggering of apoptosis following γ-radiation.
Despite the importance of Puma as a p53-dependent proapoptotic gene, a substantial fraction of the cells dying in the bone marrow must rely on DR5-signaling for efficient triggering of apoptosis, suggesting that DR5 may act upstream of PUMA for the initiation of radiation-induced apoptosis in (some) bone marrow cells. Indeed, irradiated Dr5−/−; Puma−/− mice did not show any detectable synergistic protection from cell death in any of the radio-sensitive organs investigated (Fig. 4), indicating that PUMA and DR5 trigger radiation-induced apoptosis through a common mechanism.
Loss of caspase 9 cleavage correlates with radioprotection in Puma−/− and Trp53−/− and, to a lesser extent, Dr5−/− animals.
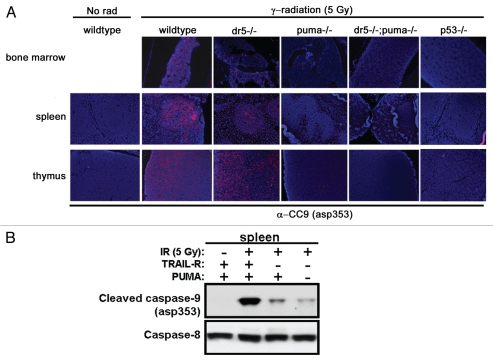
The contribution of the intrinsic pathway to caspase 9 cleavage and apoptosis in radio-sensitive tissues of the different genotypes was analyzed by immunostaining for asp353-cleaved caspase 9 (CC9) (Fig. 5). In general, the organ compartments containing caspase 9 cleavage correlated well with the TUNEL staining. In the spleen, CC9 was localized to the white pulp, a compartment that has been shown to be highly dependent on p53 for the triggering of cell death following DNA damage.30,31 Indeed, CC9 was essentially lost in the hematopoietic and lymphoid organs following loss of PUMA or p53 (Fig. 5A). However, the presence of CC9 was overall less affected following loss of DR5 in the bone marrow and spleen and very modestly affected in the Dr5-deficient thymus following ionizing irradiation. Protein gel blotting showed decreased levels of CC9 in the spleen of irradiated Dr5−/− animals; further inhibition of caspase 9 cleavage was detected in irradiated spleens from Dr5−/−; Puma−/− animals (Fig. 5B). Thus in contrast to the result measuring cell death, caspase 9 cleavage was synergistically inhibited by loss of both DR5 and PUMA, and caspase 9 cleavage only partially correlate with DNA fragmentation and apoptosis.
Figure 5.
γ-radiation-induced caspase 9 activation is attenuated in p53-, PUMA- and DR5-depleted organs. (A) Lymphoid organs from mice irradiated with 5 Gy analyzed by immunohistochemistry for murine cleaved caspase 9 (asp353) (red-cy3) showed an almost complete absence of positive cells with the loss of p53 or Puma. (B) The presence of cleaved caspase 9 was reduced following irradiation, as substantiated by protein gel blotting. Fulllength caspase 8 was used as a loading control.
Discussion
The by now well-investigated transcription factor and tumor suppressor p53 has the ability to trigger programmed cell death in normal and cancer cells alike following a range of different types of cellular stress, such as genotoxic damage. We were able to verify p53 as an important regulator of apoptosis and longterm survival in the colorectal cancer cell line HCT116 following 5FU (Fig. 1), and subsequently, treatment with chemotherapeutics also led to a p53-dependent upregulation of the expression of several PPGs (Fig. 2). Bax, Puma, Bad, Bnip3L, Fas and DR5 showed p53-dependent upregulation following ADR and 5FU in HCT116 cells, whereas we were unable to verify upregulation of Bak, Bid and caspase 6 under these conditions (Fig. 2).
The reason for the apparent redundancy in PPGs is largely unclear. The terminal nature of apoptotic cell death may require multiple layers of regulation to control appropriate initiation of apoptosis. Furthermore, the apparent redundancy of PPGs may also define a broad generalized tissue specificity, where only a select few pro-apoptotic molecules have a prominent role in each and any given tissue and cell in a context-dependent manner. Our data are in agreement with this hypothesis. Following treatment with 5-FU, DR5, Fas, Bax, Puma, Noxa and Bnip3L all contributed to caspase 3/7 cleavage in the colorectal cancer cell line HCT116, whereas we were unable to substantiate a role for Bad under these conditions (Fig. 3B). Short inhibitory RNA blockage of DR5 was most efficient in inhibiting caspase 3/7 activation (80% inhibition of caspase 3/7 cleavage relative untreated control) of all the genes tested following 5FU treatment. Indeed, previously published data documents the importance of DR5 signaling in the apoptotic response to 5FU in colorectal cancer cells.21 The finding that attenuation of Bad expression did not significantly influence caspase 3/7 cleavage was somewhat surprising, since Bad may regulate cell death in lung cancer cells following treatment with etoposide, and Bad−/− thymocytes are resistant to radiation-induced death in vitro.32,33 This suggests that Bad may have a limited role in HCT116 cells in the control of apoptosis following treatment with 5FU.
The relative synergies between the different PPGs have not been well established. In order to address this issue, we took advantage of siRNA technology to simultaneously block pairs of the PPGs in the HCT116 cells (Fig. 3). We observed a small inhibition of caspase 3/7 activity from Bax, Puma and Bnip3L following silencing of DR5 (Fig. 3C), suggesting that there is no functional overlap between the extrinsic and intrinsic pathway in HCT116 cells following treatment with 5FU. In contrast to DR5, we only observed an additive effect between Fas and Bnip3L-inhibition with respect to caspase 3/7 cleavage (Fig. 3D). One possible interpretation of these findings is that Fas employs Puma and Bax but not Bnip3L to generate signals that cleave caspase 3/7, whereas Puma and Bax act on a distinct signal transduction pathway following DR5 activation in HCT116 cells.
The in vivo role for DR5 and PUMA following ionizing radiation.
Mice lacking Dr5 and Puma are protected from apoptosis in the brain, bone marrow, spleen and thymus following ionizing irradiation.19 In vitro findings support a role for DR5 in the DNA damage response, as expression of dominant-negative DR5 or knockdown of DR5 protects different tumor cell lines, independently of exogenous TRAIL, from cell death induced by DNA damage.21,34 Puma-deficient mice are protected from a number of pro-apoptotic cellular stresses that may cause cell death in a p53-dependent or p53-independent manner. For example Pumadeficient mice have been shown to be resistant to cardiomyocyte death following ischemia reperfusion.35 Independently of p53, PUMA has been shown to promote apoptosis following stimulation with TNFα, and this is a result of increased expression of the Puma gene mediated by the p65 component of NFκB.36
Apoptosis in vivo following sub-lethal whole-body ionizing radiation were almost exclusively p53-depdent in the bone marrow, spleen, thymus and GI tract (Fig. 4). This is in agreement with what has been previously published in reference 31 and 37. Both Dr5 and Puma contributed significantly to cell death in the bone marrow, spleen and thymus. However, Puma represented a dominant gene in the bone marrow, where loss of a single allele was sufficient to block cell death following γ-radiation (Fig. 4B). Concomitant loss of Dr5 and Puma did not offer any additional protection from apoptosis following ionizing radiation to mimic that observed in p53−/− mice in the thymus and spleen, suggesting that cell death following irradiation in these organs relies on mechanisms downstream of p53 other than DR5 or PUMA.
Although identifying Bid as a connector between the extrinsic (death receptor mediated apoptosis) and intrinsic (mitochondrial apoptosis) pathway, the present model does not directly support an interaction between DR5 and PUMA in the execution of radiation-induced apoptosis. Thus, based on previously published data, anti-apoptotic synergism would have been anticipated with lack of both PUMA and DR5 following ionizing radiation, if crosstalk between the extrinsic and intrinsic pathway would have occurred exclusively through Bid. Thus, our data might be interpreted as an indication that DR5 and PUMA are redundant in the execution of radiation-induced apoptosis in mice in vivo.
In conclusion, our collective data supports cell-specific mechanisms in the triggering of p53-dependent apoptosis following DNA damage. Our in vivo data suggest a molecular mechanism exists that places pro-apoptotic members of the intrinsic pathway downstream of TRAIL death receptor signaling following DNA damage in certain cells and tissues, or, alternatively, a common pro-apoptotic molecule (e.g., Bid) downstream of both is rate limiting for the onset of apoptosis. However, additional experiments are required to reveal the precise biochemical mechanism in greater detail. Potentially, the presence of BH3-only proapoptotic molecules may be an important predictor of the tumor response to combination therapies utilizing TRAIL or anti-DR5 antibodies in concert with DNA damaging therapy. Along those lines, small inhibitory molecules targeting Bcl-2 and Bcl-XL have been shown to sensitize TRAIL-resistant glioma cells to TRAIL death receptor-mediated apoptosis.38 Thus, identifying critical molecules at the level of mitochondria may be important in elucidating targets that may enhance TRAIL death receptor agonists activity on human malignancies.
Materials and Methods
Mice.
Mice harboring targeted mutations of the Dr5 alleles19 were crossed with mice with targeted Puma alleles.16 Dr5+/−; Puma+/− mice were used to generate Dr5−/−; Puma−/−, Puma−/−, Dr5−/− and wild-type (i.e., Dr5+/+; Puma+/+) littermates. Litters were genotyped as previously described in reference 16 and 19.
Ionizing irradiation treatment.
At 4–6 weeks of age, mice received a single dose of 5 Gy whole-body γ-irradiation from 137Cs source (at a dose rate of 1.4 Gy/min), according to a University of Pennsylvania Institutional Animal Care and Use Committee (IACUC) protocol. At 8 h after the treatment, animals were euthanized. Tissues were harvested and fixed in freshly prepared 4% paraformaldehyde overnight at 4°C. The samples were washed in phosphate-buffered saline and then transferred to 70% ethanol and embedded in paraffin for histology, snapfrozen in liquid nitrogen for protein gel blot, or single-cell suspensions were prepared for flow cytometric analysis.
In vitro cell culture.
The human colorectal carcinoma cell line HCT116, osteosarcoma cell line U2OS, lung cancer cell line H460 and teratocarcinoma cell line PA-1 were purchased from ATCC. P53−/− HCT116 cells were kindly provided by B. Vogelstein (Johns Hopkins University, Baltimore, MD). HCT116 and PA-1 cells were cultured in McCoy's 5A and BME medium supplemented with 10% FCS and antibiotics in humidified 5% CO2 at 37°C, respectively.
Assessment of long-term cell viability.
To assess the long-term effect of chemotherapy, 5 × 104 cells were seeded in a well of 24-well plates in duplicate. Next day, cells were treated with 5FU (0, 0.5, 1.0, 5.0 and 10.0 µg/ml) or with Adriamycin (0, 0.05, 0.1, 0.5 and 1.0 µg/ml). The media containing the drugs were changed every 48 h, and the culture was maintained for 7 d, and then the remaining cells were fixed with 50% methanol and stained with Coomassie brilliant blue (CBB).
sub-G1 analysis.
After the indicated treatments, the cells were collected and fixed with 70% ethanol at 4°C. The samples were stained with propidium iodide (Sigma) and subjected to flow cytometric analysis using Epics Elite flow cytometer (Beckman-Coulter, Fullerton, CA). In vivo studies, the bone marrow, spleen and thymus were isolated 8 h after γ-irradiation; single cell suspensions were prepared and analyzed for sub-G1 FACS analysis.
Protein gel blot analysis.
Protein gel blotting was performed by standard methods. Apoptotic responses in in vivo studies were performed as described previously in reference 5. Briefly, snapfrozen tissues were thawed quickly and homogenized in radioimmunoprecipitation assay (RIPA) buffer (1x phosphate-buffered saline, 1% Nonidet P-40 or Igepal CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate). Homogenates were sonicated and centrifuged at 14,000 rpm for 20 min, and the protein concentration of the supernatants as determined by the Bradford method (Bio-Rad). The samples were separated on a Tris-Glycine gel (Invitrogen) under denaturing conditions. Proteins were electro-blotted onto a polyvinylidene difluoride membrane. After incubation with the indicated antibodies, the proteins were detected using ECL Protein gel blotting Detection kit (GE Healthcare, Piscataway, NJ).
Anti-Bax, anti-Bcl2 and anti-DR4 antibodies were purchased from Becton Dickinson (Franklin Lakes, NJ). Anti-DR5 antibody was purchased from Sigma (St. Louis, MO). Anti-Bad, anti-Bid, anti-survivin, anti-mouse cleaved caspase 9, anti-mouse caspase 8, anti-caspase 6 antibodies were purchased from Cell Signaling (Boston, MA). Anti-p53 (DO1), anti-Fas and anti-Bnip3L antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Bak, anti-Noxa, anti-WAF1 (p21) and anti-Puma antibodies were purchased from Calbiochem (San Diego, CA). Following primary antibody incubation, membranes were incubated with secondary horseradish peroxidase-conjugated antibodies (1:4,000) (Pierce, Rockford, IL) and subsequently detected by the enhanced chemiluminescence method (Amersham/GE Healthcare Biosciences, Piscataway, NJ).
RT-PCR analysis.
Cells were washed, collected, and total RNA was prepared using Trizol LS Reagent (Invitrogen) according to the manufacturer's instruction. One µg total of RNA was reverse-transcribed using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA) according to the manufacture's instruction. For amplification of p53AIP1 and their splice variants, 5′-ATG GGA TCT TCC TCT GAG GC-3′ was used as a common forward primer. 5′-TCA CTG CAA CCT CAA CGG TG-3′ was used as a reverse primer. 5′-TTA CTG CAC TGT CAG GAT CC-3′ and 5′-TCA GTT CCC AGC TCT GTC CA-3′ were used as reverse primers for the β-isoform and γ-isoform, respectively. PCR reaction consists of 95°C for 30 sec, 56°C for 1 min and 72°C for 1 min and amplified from 25 to 35 cycles. The PCR products were separated by 2% agarose gel every five cycles for smi quantification, and the bands were detected using Molecular Imager Gel Documentation System (Bio-Rad Laboratories, Hercules, CA).
Gene silencing using siRNA.
For transient silencing of the p53 target genes, siRNAs for PUMA, DR5 and FAS as well as non-silencing control were purchased from Santa Cruz Biotechnology. 5′-AGG AUG AGG AUG GUA CGU G-3′ and 5′-AUA UUC AAG AUG GCU GCC C-3′ were used to silence BNIP3L and BAD , respectively. RNA TransPass RI Transfection Reagent (New England BioLabs) was used for the transfection of siRNA according to the manufacturer's instruction.
Measurement of caspase 3 and -7 activities.
7,500 cells HCT116 cells were seeded in a 96-well plates on day 1. siRNA-transfection was performed on day 2. Adriamycin (1 µg/ml) or 5FU (20 µg/ml) treatment started on day 3. Caspase 3/7 activities were measured 24 h after the treatment using Caspase Glo 3/7 Assay (Promega, Madison, WI) according to the manufacture's instructions. The luminescent signal was detected, visualized and measured using Xenogen In vivo Imaging System (Xenogen, Hopkinton, MA). After gene silencing and the drug treatment, all wells were checked under microscopy to ensure that there were no differences in cell numbers between wells.
Immunohistochemistry.
Immunohistochemistry was performed as previously described in references 5, 19 and 39. Briefly, the bone marrow, spleen, thymus, small intestine and colon were collected and fixed in 4% paraformaldehyde and embedded in paraffin. Tissue-paraffin blocks were cut in 5 µm. Paraffin sections were dewaxed and immunostained with primary antibodies and the secondary antibodies conjugated with Cy3 (Jackson Immunochemicals). Anti-cleaved caspase 9 (asp353, mouse specific, 1:100) antibody was purchased from Cell Signaling. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed using the Apoptag plus FITC kit according to the manufacturer's instructions (Chemicon International). Sections were counterstained with 4′-6-diamidino-2-phenylindole (DAPI) or propidium iodide (PI) and evaluated under a fluorescence microscope.
Supplementary Material
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22:7486–7495. doi: 10.1038/sj.onc.1206949. [DOI] [PubMed] [Google Scholar]
- 4.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 5.Kuribayashi K, Finnberg N, El-Deiry WS. Studying p53-dependent cell death in vitro and in vivo. Methods Enzymol. 2008;446:159–173. doi: 10.1016/S0076-6879(08)01609-1. [DOI] [PubMed] [Google Scholar]
- 6.Kuribayashi K, El-Deiry WS. Regulation of programmed cell death by the p53 pathway. Adv Exp Med Biol. 2008;615:201–221. doi: 10.1007/978-1-4020-6554-5_10. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Mehew JW, Heckman CA, Arcinas M, Boxer LM. Negative regulation of bcl-2 expression by p53 in hematopoietic cells. Oncogene. 2001;20:240–251. doi: 10.1038/sj.onc.1204067. [DOI] [PubMed] [Google Scholar]
- 9.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 10.Komarova EA, Chernov MV, Franks R, Wang K, Armin G, Zelnick CR, et al. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 1997;16:1391–1400. doi: 10.1093/emboj/16.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merritt AJ, Potten CS, Kemp CJ, Hickman JA, Balmain A, Lane DP, et al. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 1994;54:614–617. [PubMed] [Google Scholar]
- 12.Merritt AJ, Allen TD, Potten CS, Hickman JA. Apoptosis in small intestinal epithelial from p53-null mice: evidence for a delayed, p53-independent G2/M-associated cell death after gamma-irradiation. Oncogene. 1997;14:2759–2766. doi: 10.1038/sj.onc.1201126. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard DM, Potten CS, Hickman JA. The relationships between p53-dependent apoptosis, inhibition of proliferation and 5-fluorouracil-induced histopathology in murine intestinal epithelia. Cancer Res. 1998;58:5453–5465. [PubMed] [Google Scholar]
- 14.Anderson CN, Tolkovsky AM. A role for MAPK/ERK in sympathetic neuron survival: protection against a p53-dependent, JNK-independent induction of apoptosis by cytosine arabinoside. J Neurosci. 1999;19:664–673. doi: 10.1523/JNEUROSCI.19-02-00664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/S1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 17.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/S1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 19.Finnberg N, Gruber JJ, Fei P, Rudolph D, Bric A, Kim SH, et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol Cell Biol. 2005;25:2000–2013. doi: 10.1128/MCB.25.5.2000-13.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forand A, Bernardino-Sgherri J. A critical role of PUMA in maintenance of genomic integrity of murine spermatogonial stem cell precursors after genotoxic stress. Cell Res. 2009;19:1018–1030. doi: 10.1038/cr.2009.50. [DOI] [PubMed] [Google Scholar]
- 21.Longley DB, Wilson TR, McEwan M, Allen WL, McDermott U, Galligan L, et al. c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene. 2006;25:838–848. doi: 10.1038/sj.onc.1209122. [DOI] [PubMed] [Google Scholar]
- 22.Seitz SJ, Schleithoff ES, Koch A, Schuster A, Teufel A, Staib F, et al. Chemotherapy-induced apoptosis in hepatocellular carcinoma involves the p53 family and is mediated via the extrinsic and the intrinsic pathway. Int J Cancer. 2010;126:2049–2066. doi: 10.1002/ijc.24861. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, El-Deiry WS. Inducible silencing of KILLER/DR5 in vivo promotes bioluminescent colon tumor xenograft growth and confers resistance to chemotherapeutic agent 5-fluorouracil. Cancer Res. 2004;64:6666–6672. doi: 10.1158/0008-5472.CAN-04-1734. [DOI] [PubMed] [Google Scholar]
- 24.Corazza N, Jakob S, Schaer C, Frese S, Keogh A, Stroka D, et al. TRAIL receptor-mediated JNK activation and Bim phosphorylation critically regulate Fas-mediated liver damage and lethality. J Clin Invest. 2006;116:2493–2499. doi: 10.1172/JCI27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 26.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/S0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 27.Wu GS, Burns TF, McDonald ER, 3rd, Jiang W, Meng R, Krantz ID, et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 28.Kuribayashi K, Krigsfeld G, Wang W, Xu J, Mayes PA, Dicker DT, et al. TNFSF10 (TRAIL), a p53 target gene that mediates p53-dependent cell death. Cancer Biol Ther. 2008;7:2034–2038. doi: 10.4161/cbt.7.12.7460. [DOI] [PubMed] [Google Scholar]
- 29.Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- 31.Komarova EA, Christov K, Faerman AI, Gudkov AV. Different impact of p53 and p21 on the radiation response of mouse tissues. Oncogene. 2000;19:3791–3798. doi: 10.1038/sj.onc.1203717. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally upregulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol Cell Biol. 2006;26:9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9324–9329. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong B, Almasan A. Apo2 ligand/TNF-related apoptosis-inducing ligand and death receptor 5 mediate the apoptotic signaling induced by ionizing radiation in leukemic cells. Cancer Res. 2000;60:5754–5760. [PubMed] [Google Scholar]
- 35.Toth A, Jeffers JR, Nickson P, Min JY, Morgan JP, Zambetti GP, et al. Targeted deletion of Puma attenuates cardiomyocyte death and improves cardiac function during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2006;291:52–60. doi: 10.1152/ajpheart.01046.2005. [DOI] [PubMed] [Google Scholar]
- 36.Wang P, Qiu W, Dudgeon C, Liu H, Huang C, Zambetti GP, et al. PUMA is directly activated by NFkappaB and contributes to TNFalpha-induced apoptosis. Cell Death Differ. 2009;16:1192–1202. doi: 10.1038/cdd.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 38.Hetschko H, Voss V, Horn S, Seifert V, Prehn JH, Kogel D. Pharmacological inhibition of Bcl-2 family members reactivates TRAIL-induced apoptosis in malignant glioma. J Neurooncol. 2008;86:265–272. doi: 10.1007/s11060-007-9472-6. [DOI] [PubMed] [Google Scholar]
- 39.Finnberg N, Klein-Szanto AJ, El-Deiry WS. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J Clin Invest. 2008;118:111–123. doi: 10.1172/JCI29900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.