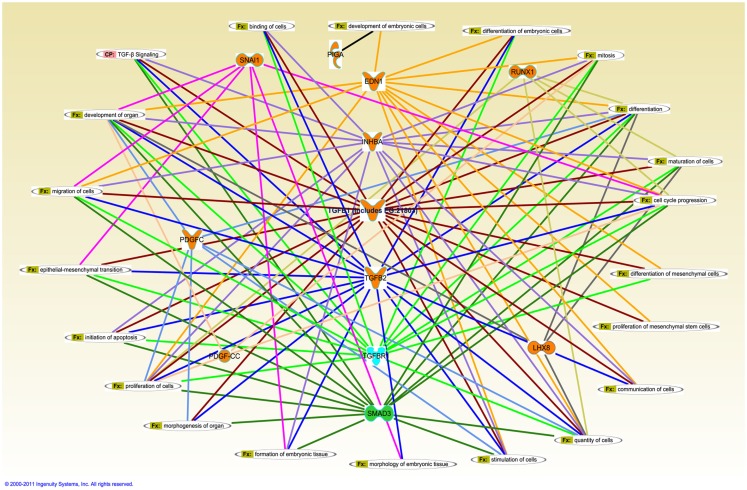
Figure 6.
Merged interaction network of cleft palate susceptibility genes and biofunctions. Affymetrix probe identifiers and fold-values were uploaded to IPA and each identifier was mapped to its corresponding gene object in the IKB. Interactions were then queried between these gene objects and all other gene objects stored within IPA to generate a set of direct interaction networks that were merged. Cellular functions that directly interact with differentially expressed putative candidate genes for cleft palate in response to TGFβ treatment of HEPM cells were included in the network. The 11 transcripts meeting the 0.05 criteria of FDR in response to TGFβ treatment were analyzed for the direct interaction network using IPA. Upregulated genes (EDN1, INHBA, TGFβ1, TGFβ2, RUNX1, LHX8, SNAI1, PDGFC, and PIGA) were highlighted with orange and downregulated gene (SMAD3) was highlighted with green. TGFβR1 was upregulated with TGFβ1 treatment and downregulated with TGFβ2 treatment, thus highlighted with cyan. Fx, cellular function; CP, canonical pathway.