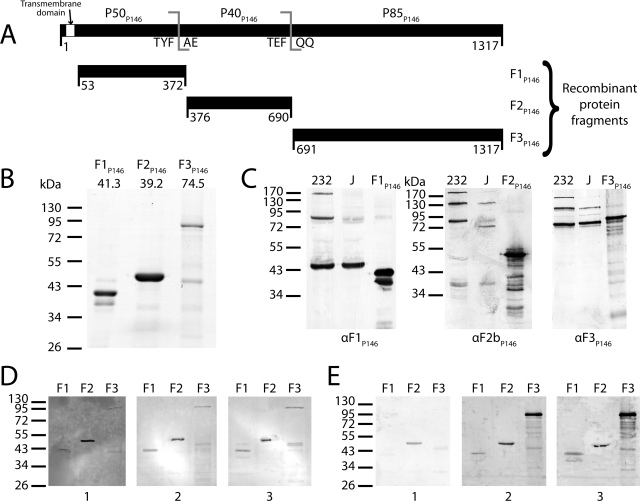
FIG 2 .
Cloning of p146 and immunoblot analysis of its cleavage fragments. (A) p146 (3954 bp encoding 1317 residues) was cloned from M. hyopneumoniae strain 232 homolog mhp684 in three fragments (F1P146/232 to F3P146/232) closely matching native cleavage as observed by mass spectrometry. A hydrophobic transmembrane domain predicted by TMHMM (53 residues) was removed from the N terminus of F1P146/232. All in-frame TGA codons were mutated to TGG using overlap extension PCR. (B) Coomassie blue-stained SDS-PAGE of the three P146 recombinant proteins. The predicted mass (in kilodaltons) is shown above each gel lane. Recombinant protein F2P146/232 runs at a higher molecular mass than predicted. (C) Immunoblots of recombinant proteins (F1P146/232 to F3P146/232) and whole-cell lysates from M. hyopneumoniae strains 232 and J, probed with anti-F1P146 (αF1P146) and anti-F3P146 (αF3P146) sera. Sera labeled αF1P146 and αF3P146 were extracted from New Zealand White rabbits challenged with recombinants F1P146/232 and F3P146/232; anti-F2bP146 (αF2bP146) serum was generated in the same manner from a recombinant protein representing residues 586 to 690 of P146. Minor bands present in recombinant protein lanes (F1P146/232 to F3P146/232) may be either products of recombinant protein degradation or truncated proteins resulting from mRNA instability. (D) Immunoblots of recombinant proteins (F1P146/232 to F3P146/232) probed with porcine serum sourced from a high-health-status herd (blot 1) and sera from convalescent pigs (blots 2 and 3). (E) Immunoblots of recombinant proteins (F1P146/232 to F3P146/232) probed with sera obtained from a single pig prior to treatment (blot 1), after vaccination with the commercial bacterin vaccine Suvaxyn (blot 2), or 6 weeks after challenge with M. hyopneumoniae strain Hillcrest (blot 3).