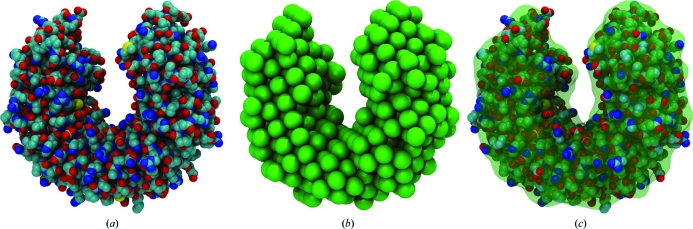
Figure 2.
Real-space fitting and visualization of SAXS bead models with Situs (Wriggers & Chacón, 2001b ▶; Wriggers, 2010 ▶). (a) Atomic structure of ribonuclease inhibitor (PDB entry 1bnh; Kobe & Deisenhofer, 1996 ▶). (b) Coarse-grained bead model (393 beads, 3 Å radius, hexagonal close-packed) generated from (a) with the Situs tool pdb2sax. (c) Atomic structure fitted to the bead model using collage. To show the embedded structure, the bead model is rendered as a transparent envelope. The envelope is the half-maximum isosurface of a density map created from the beads with pdb2vol using Gaussian kernel convolution with (half-maximum) kernel radius 3 Å. The images were rendered with Tachyon ray tracing using VMD (Humphrey et al., 1996 ▶). For an updated review of the complete SAXS workflow, see Wriggers (2010 ▶) and the SAXS tutorial at http://situs.biomachina.org.