Background: CRL4Cdt2 requires that a substrate bind to proliferating cell nuclear antigen (PCNA) on DNA prior to ligase recruitment, but the precise role of PCNA is unclear.
Results: A specific PCNA residue is required for destruction of CRL4Cdt2 substrates.
Conclusion: CRL4Cdt2 recognizes a composite surface composed of PCNA and substrate residues.
Significance: This is the first ubiquitin ligase whose substrate recognition requires creation of a bipartite substrate surface.
Keywords: Cell cycle, DNA repair, DNA replication, E3 ubiquitin ligase, Protein-protein interactions, CRL4, Cdt2, Cul4, Degron, PCNA
Abstract
The E3 ubiquitin ligase Cullin-ring ligase 4-Cdt2 (CRL4Cdt2) is emerging as an important cell cycle regulator that targets numerous proteins for destruction in S phase and after DNA damage, including Cdt1, p21, and Set8. CRL4Cdt2 substrates contain a “PIP degron,” which consists of a canonical proliferating cell nuclear antigen (PCNA) interaction motif (PIP box) and an adjacent basic amino acid. Substrates use their PIP box to form a binary complex with PCNA on chromatin and the basic residue to recruit CRL4Cdt2 for substrate ubiquitylation. Using Xenopus egg extracts, we identify an acidic residue in PCNA that is essential to support destruction of all CRL4Cdt2 substrates. This PCNA residue, which adjoins the basic amino acid of the bound PIP degron, is dispensable for substrate binding to PCNA but essential for CRL4Cdt2 recruitment to chromatin. Our data show that the interaction of CRL4Cdt2 with substrates requires molecular determinants not only in the substrate degron but also on PCNA. The results illustrate a potentially general mechanism by which E3 ligases can couple ubiquitylation to the formation of protein-protein interactions.
Introduction
Eukaryotic cells contain hundreds of E3 ubiquitin ligases that each ubiquitylate one or more target proteins, modulating their activity or marking them for destruction by the 26 S proteasome (1). Several of the best-studied E3 ligases regulate cell cycle progression. For example, the Skp2-containing Cullin ring ligase 1 (CRL1Skp2 also known as SCFSkp2)2 is composed of a Cul1 scaffold, a Skp1 adaptor, the Skp2 substrate receptor, and Rbx1, which interacts with a ubiquitin-conjugating enzyme. Skp2 binds directly to substrates via a “phosphodegron,” a short peptide motif on the substrate whose phosphorylation by cyclin-dependent kinases promotes its interaction with the leucine-rich repeat motif of Skp2 (2, 3). In late G1 phase, when CDK activity rises, CRL1Skp2 targets the CDK inhibitor p27 for destruction, promoting S phase entry. Another cell cycle-regulated ubiquitin ligase is the anaphase-promoting complex (APCCdc20). This multisubunit enzyme targets a number of factors for destruction, including Cyclin B and securin (4). In this case, the ligase itself is phosphorylated by mitotic CDKs, leading to substrate-ligase interactions.
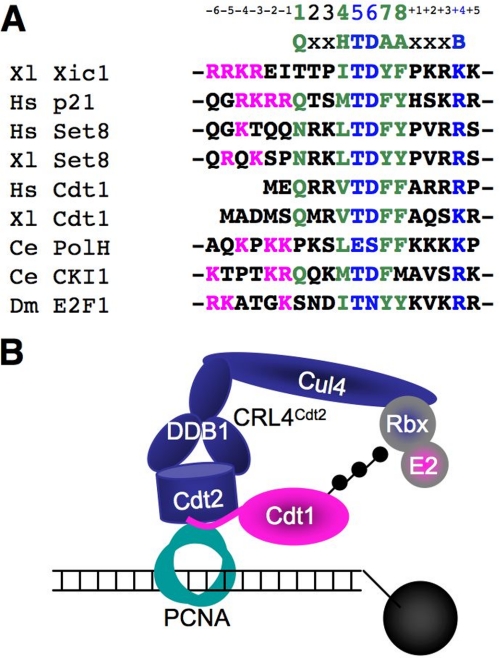
Recently, an unusual ubiquitin ligase called CRL4Cdt2 has been characterized, which promotes the ubiquitylation of several proteins in S phase and after DNA damage (5, 6). CRL4Cdt2 is composed of a Cul4 scaffold, a Ddb1 adaptor, and Cdt2, the putative substrate receptor. In vertebrates, CRL4Cdt2 targets the licensing factor Cdt1 (6–9), the CDK inhibitor p21 (Xic1 in frogs) (10–13), and the histone methyltransferase Set8 (14–18) for proteolysis in S phase. In all three cases, destruction appears to contribute to the block to re-replication. Set8 destruction also promotes transcription and prevents premature chromatin compaction (14, 15, 18). CRL4Cdt2 targets the transcription factor E2F in flies (to shut off G1 transcription in S phase and to regulate endocycles) (19, 20), the translesion DNA polymerase η in worms (perhaps to restrict access of this mutagenic polymerase to undamaged DNA) (21), and the ribonucleotide reductase inhibitor Spd1 in fission yeast (to activate nucleotide synthesis in S phase and after DNA damage) (22). Other substrates of CRL4Cdt2 are likely to emerge.
The activity of CRL4Cdt2 is coupled to DNA replication and damage via PCNA, a homotrimeric, ring-shaped molecule that encircles DNA. Given its topological embrace of DNA, PCNA tethers to DNA any proteins with which it interacts, including DNA polymerases, DNA ligases, chromatin-remodeling factors, and numerous other proteins involved in DNA replication and repair (23). Most proteins that bind PCNA do so via an eight-amino acid motif called a PIP box (see Fig. 1A, green amino acids). The aromatic and hydrophobic residues in the PIP box interact with a hydrophobic pocket underlying the interdomain connector loop (IDCL) (23–26). CRL4Cdt2 substrates contain a PIP box, through which they bind to chromatin-bound PCNA (PCNAchromatin) at sites of DNA damage or at the replication fork. Most CRL4Cdt2 substrates also contain a TD motif at positions 5 and 6 of the PIP box (Fig. 1A, blue amino acids), which confers especially high affinity binding to PCNA (27, 28). However, a PIP box and TD motif are not sufficient for CRL4Cdt2 activity. All substrates also contain a basic residue four amino acids downstream of the PIP box (the “B+4” residue). When this residue is mutated to alanine in Xenopus Cdt1, the resulting protein binds normally to PCNA, but CRL4Cdt2 is not recruited to the Cdt1·PCNA complex (27). These data explain why most PIP box proteins, such as DNA polymerases, which lack the B+4 residue, are not destroyed. In summary, the above data indicate that, during CRL4Cdt2-mediated proteolysis, a substrate docks onto PCNAchromatin, leading to recruitment of CRL4Cdt2, followed by ubiquitin transfer (see Fig. 1B). Notably, an alternative model has recently been proposed in which CRL4Cdt2 binds PCNA independently of a substrate, and in which the PCNA and Cdt2 binding regions within substrates can be separated (11). Thus, there is significant disagreement over the role of PCNA in promoting substrate recognition by CRL4Cdt2.
FIGURE 1.
PCNA-dependent recognition of PIP degrons by CRL4Cdt2. A, sequence alignment of CRL4Cdt2 substrate PIP degrons. Canonical PIP box residues are shown in green. H stands for a hydrophobic residue (Ile, Leu, Val, or Met) and A stands for an aromatic residue (Phe or Tyr). The PIP degron residues are shown in blue, B stands for any basic residue (Lys or Arg). The upstream basic residues are shown in pink. B, model of substrate recognition on PCNA by CRL4Cdt2 on the immobilized 1-kb DNA template.
In this report, we investigate the mechanism by which CRL4Cdt2 interacts with its substrates to trigger proteolytic degradation. First, we provide evidence against the recent proposal that CRL4Cdt2 and its substrates dock onto PCNA independently of one another (11), thereby affirming that CRL4Cdt2 is initially recruited to a PIP degron·PCNA complex. We then addressed whether the sole function of PCNA is to position the substrate's PIP degron for binding to CRL4Cdt2 (indirect role) or if a specific surface of PCNA is required, together with the PIP degron, to recruit CRL4Cdt2 (direct role). In support of the latter model, we identify an acidic residue (Asp-122) on the surface of PCNA that is essential for CRL4Cdt2 activity. Importantly, this residue is not necessary for PIP box binding to PCNA but is essential for CRL4Cdt2 recruitment to the PCNA·PIP degron complex on chromatin. Asp-122 is also essential for CRL4Cdt2 activity in fission yeast. Our findings support the idea that CRL4Cdt2 recognizes the PCNA·PIP degron interface by making direct contacts with residues in both polypeptides. This mechanism suggests new possibilities for the regulation of proteolysis.
EXPERIMENTAL PROCEDURES
Xic1 in Vitro Transcription and Translation
pCS2+/Xic1 (11) was in vitro translated according to the manufacturer's protocol using TnT® SP6 Quick Coupled Transcription Translation kit from Promega (Madison, WI). In each reaction 125 ng of DNA was used per 10 μl of TNT® Quick Master Mix (Promega), and reactions were scaled according to the amount of Xic1 needed.
Egg Extract and Immunological Methods
High speed supernatant (HSS), low speed supernatant (LSS) (29–31), and chromatin spin-down assays (32) were performed as described. We used previously described antibodies against Cdt1 (32), Ddb1 (33), RPA (30), Cdt2 (7), GST (New England Biolabs), M2 and Rabbit FLAG (Sigma), Xenopus PCNA (34), and PCNA (Santa Cruz Biotechnology, sc-056).
Depletion of Xenopus PCNA
The polyclonal PCNA antibody used for PCNA depletion was generated as described previously (34). To deplete PCNA from HSS, three rounds of depletion were performed using 3 μl of PCNA antibody per 1 μl of rProtein A-Sepharose FastFlow resin (Amersham Biosciences). To deplete PCNA from LSS, two rounds of depletion were performed. The antibody and resin were pre-bound, and 0.2 volume of resin was used per microliter of egg extract.
Cloning and Protein Purification for Xenopus Egg Extract Experiments
Recombinant Xenopus Cdt11–243-3xNLS-GST-FLAG (27), human PCNA (27), and GST-FLAG-tagged human Set8 (15) were previously described. Mutations in human PCNA were generated using a QuikChange mutagenesis kit (Clontech) and the following primers and reverse compliments: D120A, GACTATGAAATGAAGTTGATGGCTTTAGATGTTGAACAACTTGG; D122A, GAAATGAAGTTGATGGATTTAGCTGTTGAACAACTTGGA; E124A, ATGGATTTAGATGTTGCACAACTTGGAATTCCAGAACAG; D122A/E124A, GATGGATTTAGCTGTTGCTCAACTTGGAATTCC; and D122K/E124R, GATGGATTTACGTGGTAAACAACTTGGAATTCC.
MMS DNA Preparation and Bead Spin-down Assay
Methylated DNA was generated as described previously (35). A biotinylated 1-kb PCR product was generated, treated with methyl methane sulfonate (MMS), and coupled to M-280 streptavidin Dynabeads (Invitrogen) as previously described (27). To spin down MMS-DNA beads, we used a modification of our standard chromatin spin-down protocol (33), in which the egg lysis buffer (10 mm HEPES (pH 7.7), 250 mm sucrose, 50 mm KCl, 2.5 mm MgCl2) wash step was supplemented with 0.6% Triton X-100.
Fission Yeast General Methods
Strains used in this study are listed in supplemental Table S1. Standard genetic methods and flow cytometry were used as described previously (36, 37).
PCNA Mutagenesis in Fission Yeast
To construct the pcn1-D122A mutant strain 2640, two partially complementary fragments for the mutation (NsiI-fragment and BamHI-fragment) were amplified using primers containing the mutation and an NsiI or BamHI restriction site; subsequently, the fragments were annealed and amplified. After DNA purification, the fragment was cloned into NsiI- and BamHI-digested pSMH and integrated at the pcn1+ locus after linearizing with XhoI (in the strain 2069). Hygromycin-resistant transformants were selected, and the correct integration of the plasmid was checked with the primers 619 and 979. The plasmid and all the strains constructed were checked by sequencing (primer 978). Primers used are shown in supplemental Table S2.
Cell Cycle Synchronization and UV Irradiation Experiments
Cells were arrested in G1 phase in Edinburgh minimal medium (65) lacking NH4Cl for 16 h at 25 °C (38), then released into the cell cycle in rich yeast extract and supplements medium (65) at 32 °C. nda3–311 strains were grown at 32 °C in rich medium and arrested in M phase by incubation for 4 h at 20 °C. For UV exposure, cells were resuspended in water and irradiated with 100 J/m2 of 254 nm UV light in a 6-mm deep stirred suspension at 20 °C.
Protein Analysis of Fission Yeast Samples
Protein extracts were made by TCA extraction and analyzed by Western blotting as described previously (39). Tandam affinity purification (TAP)-tagged Cdt1 was detected with peroxidase·anti-peroxidase-soluble complex (P1291, Sigma), and α-tubulin was used as loading control and detected with antibody T5168 (Sigma).
RESULTS
To study the mechanism of CRL4Cdt2-mediated proteolysis, we used two approaches that involve different types of Xenopus egg extracts (summarized in supplemental Fig. S1). First, DNA that had been treated with MMS to induce methylation damage was added to an HSS of egg cytoplasm. Nucleotide excision repair of the methylation damage involves a PCNA-dependent gap-filling step that promotes destruction of endogenous Cdt1 as well as other substrates by CRL4Cdt2 (7, 15). Second, sperm chromatin was added to an LSS of egg cytoplasm. Upon nuclear-envelope assembly around the sperm, chromosomal DNA replication initiates, leading to PCNA-dependent, CRL4Cdt2-mediated proteolysis (7, 33). Most experiments were performed using DNA damage-induced destruction in HSS, but key conclusions were confirmed using the S phase pathway in LSS.
The B+4 Residue Can Be Partially Compensated for in Xic1 by Residues Upstream of the PIP Box
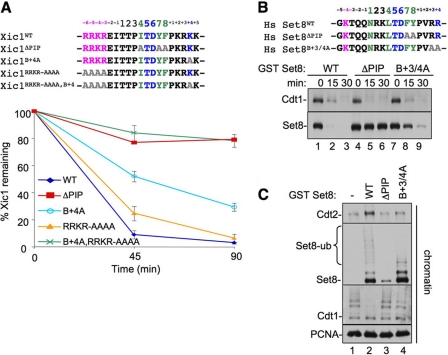
The B+4 residue is conserved in all CRL4Cdt2 substrates (Fig. 1A), and it is known to be essential for Cdt1 destruction (27, 28). We wanted to know whether B+4 is also important for destruction of other substrates. We therefore examined Xic1 and Set8. When added to Xenopus egg extracts, Xic1, a Xenopus CDK inhibitor, is destroyed in a manner that requires its PIP box, chromatin-loaded PCNA, and Cdt2 (11, 40–42). However, in contrast to Cdt1B+4A, which was completely stable (27), Xic1B+4A was still destroyed, albeit more slowly than Xic1WT (Fig. 2A, bottom, light blue trace). This residual destruction of Xic1B+4A was PCNA-dependent (data not shown). We next tested the histone H4 lysine 20 methyltransferase Set8, which is also destroyed during S phase and after DNA damage in human cells or when added to Xenopus egg extracts (14–18). Like Xic1B+4A, Set8B+3/4A was destroyed slowly (Fig. 2B, lanes 7–9; note that in Set8B+3/4A, the B+3 residue was also mutated to alanine in case of charge redundancy), and the residual destruction was still PCNA-dependent (see below). Similar to Cdt1B+4A (27), Set8B+3/4A bound normally to PCNAchromatin, and its slow destruction was due to reduced recruitment of CRL4Cdt2 to chromatin, resulting in reduced ubiquitylation of Set8B+3/4A (Fig. 2C). In summary, as seen in Cdt1, the B+4 residue in Xic1 and Set8 is important for efficient destruction due to a role in CRL4Cdt2 recruitment, although it is less crucial for destruction of the latter substrates.
FIGURE 2.
The B+4 residue can be partially compensated for by residues upstream of the PIP box in Xic1 and Set8. In A: Top, sequence comparison of Xic1 PIP degron with upstream basic residues and the various mutants examined. Bottom, graph showing the percentage of in vitro translated 35S-labeled Xic1 remaining after it was added to HSS in the presence of 5 ng/μl MMS-damaged plasmid. Reactions were stopped at the indicated time points, and the amount of Xic1 remaining was quantified by autoradiography. Results from three independent experiments were averaged and graphed. Bars represent the standard error of the mean. In B: Top, sequence alignment of Set8 PIP degron with upstream residues and the various mutants examined. Bottom, HSS was supplemented with 5 ng/μl MMS plasmid and 50 nm human Set8WT, Set8ΔPIP, or Set8B+3/4A. At the indicated times, samples were blotted for endogenous Cdt1 or Set8. C, HSS was supplemented with immobilized 1-kb MMS DNA and 2 mg/ml methyl ubiquitin. Buffer and 50 nm Set8WT, Set8ΔPIP, or Set8B+3/4A was also added, and after 10 min, chromatin was recovered from the extract and washed, and the indicated proteins were visualized by Western blotting.
Given the different effects of the B+4A mutation on different CRL4Cdt2 substrates, we re-examined their sequences. Notably, the PIP box of Cdt1 is located at the extreme amino terminus, whereas the PIP boxes of p21, Xic1, and Set8 are internal or C-terminal to these proteins. Additionally, the latter substrates contain one or more basic amino acids immediately upstream of the PIP box (Fig. 1A, pink) (27). We speculated that these basic residues might contribute to destruction in the absence of the B+4 residue. To test this hypothesis, we mutated the basic residues N-terminal of the PIP box to alanines in Xic1, yielding Xic1RRKR/AAAA (Fig. 2A, top). Mutation of these residues alone had little or no effect on Xic1 destruction (Fig. 2A, yellow trace). However, when combined with the B+4 mutant, these mutations rendered the protein completely stable, similar to Xic1ΔPIP (Fig. 2A, green trace). Together, these data show that, in a CRL4Cdt2 substrate where the PIP degron is not located at the extreme N terminus, additional basic residues located just upstream of the PIP box can contribute to CRL4Cdt2-mediated destruction, but they are only essential in the absence of the B+4 mutant. The upstream basic residues likely promote substrate destruction in a manner that is distinct from the downstream basic residues, including B+4.
The PCNA and CRL4Cdt2 Binding Motifs of Substrates Cannot Be Separated
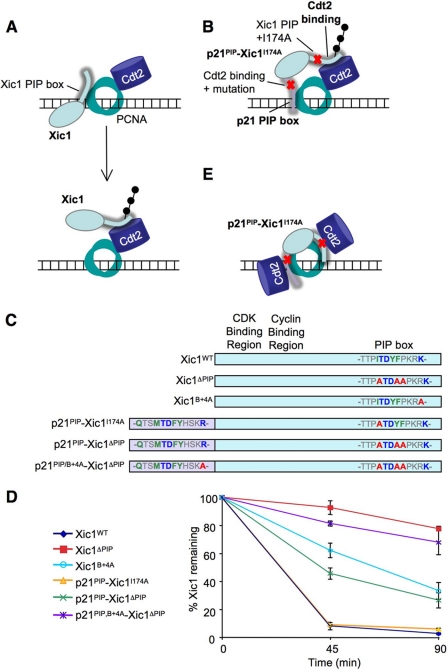
Recently, Yew and colleagues proposed a new model for substrate recognition by CRL4Cdt2 in which the substrate and CRL4Cdt2 initially bind independently to PCNA before coming together in a complex (Fig. 3A). This model was based on two considerations. First, they showed that the C terminus of Cdt2 can bind to PCNA independently of substrate (11). However, our previous data established that Cdt2 does not bind to PCNA on chromatin in the absence of substrate (7, 27), demonstrating that a Cdt2-PCNA interaction is insufficient to support chromatin recruitment of the ligase. Second, Yew and colleagues proposed that, within the primary amino acid sequence of Xic1, PIP box and Cdt2 binding motifs can be widely separated and still promote destruction (11). Their conclusion, which would further indicate that CRL4Cdt2 does not bind PCNA in the context of substrate, was based on the following experiment. The p21 PIP degron, in which a proposed Cdt2-binding region had been deleted, was fused onto the N terminus of Xic1, in which the endogenous PIP box had been compromised through mutation of Ile-174 to alanine, yielding p21PIP-Xic1I174A (Fig. 3, B and C; originally named NPIP2- Xic1I174A in Ref. 11). This construct was destroyed normally in egg extracts (11) even though it was thought to contain well separated PCNA and Cdt2 interaction motifs, as illustrated in Fig. 3B. We repeated this experiment and obtained the same result (Fig. 3D, yellow trace). However, this experiment has two caveats. First, the B+4 residue of the added p21 PIP degron was not mutated, likely still allowing binding to Cdt2. Second, only one residue (Ile-174) in the endogenous Xic1 PIP box was mutated. Thus, the mutated Xic1 PIP box and the added p21 PIP box might bind cooperatively via interactions with two PCNA subunits (Fig. 3E). In this case, each PIP box might be able to recruit some CRL4Cdt2 and thus promote destruction via the mechanism we previously proposed (Fig. 3E) (27).
FIGURE 3.
The Cdt2 and PCNA binding motifs of Xic1 cannot be separated. A, model of CRL4Cdt2-mediated destruction of Xic1, in which Xic1 and Cdt2 both dock onto PCNA separately (top) and then come together for ubiquitin transfer (bottom). Adapted from Ref. 11. B, model for destruction of p21PIP-Xic1I174A using separable Cdt2 and PCNA binding motifs. Yew and colleagues fused residues 135–164 of p21 containing the PIP box to the N terminus of Xic1 to create a p21 PIP box-Xic1 fusion protein (11). In this construct one canonical PIP box residue in Xic1 (Ile-174) was mutated to an alanine to inhibit PCNA binding, and six residues (156–161) just past the B+4 residue of p21 (residues +5 to +10) were deleted to inhibit the Cdt2-binding region proposed by Yew and colleges. C, scheme of Xic1 and p21PIP-Xic1 mutations used in our experiments. D, graph showing the percentage of 35S-labeled Xic1 mutants remaining when incubated with HSS and 5 ng/μl MMS-damaged plasmid at the indicated times. Results from three independent experiments were averaged and graphed. Bars represent the standard error of the mean. E, our model for CRL4Cdt2-mediated destruction of p21PIP-Xic1I174A binding to two PCNA monomers.
To test whether the destruction of p21PIP-Xic1I174A involved residual binding of the Xic1 PIP box to PCNA, we mutated all three Xic1 PIP box residues, creating p21PIP-Xic1I174A,Y177A,F178A (Fig. 3C; abbreviated as p21PIP-Xic1ΔPIP). Unlike p21PIP-Xic1I174A, p21PIP-Xic1ΔPIP was destroyed much less efficiently than Xic1WT (Fig. 3D, green line). From these data we conclude that ∼40% of p21PIP-Xic1I174A destruction can be attributed to residual binding of the Xic1 PIP box to PCNA. When the B+4 residue of the added p21 PIP box was also mutated to alanine, the resulting protein, p21PIP/B+4A-Xic1ΔPIP (Fig. 3C), was destroyed at background levels (Fig. 3D, purple line), indicating that the p21 PIP box retained Cdt2 binding capacity. Thus, the original p21PIP-Xic1I174A construct did not contain adequately separated PCNA and Cdt2-binding functions. When these domains are effectively separated, the resulting fusion protein is not destroyed. Therefore, there is no evidence that these domains can function separately.
Identification of a PCNA Residue That Is Essential for CRL4Cdt2-mediated Destruction
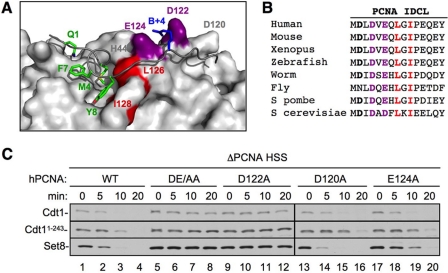
We next wanted to further characterize what role PCNA plays in the recognition of the PIP degron by CRL4Cdt2. Specifically, we wished to distinguish between direct and indirect roles for PCNA. In the “indirect” model, CRL4Cdt2 only contacts residues in the substrate's PIP degron, and the sole function of PCNA is to position the degron for recognition by CRL4Cdt2. In the “direct” model, CRL4Cdt2 recognizes a composite surface created by the two proteins. The second model predicts that substrate recognition by CRL4Cdt2 requires residues in PCNA that do not influence PIP degron binding. To look for such residues, we examined the co-crystal structure of PCNA with the PIP degron of p21 (Fig. 4A) (24). Interestingly, the structure shows that two conserved acidic residues, Asp-122 and Glu-124 on the IDCL of PCNA, cradle the B+4 residue of p21 (Fig. 4, A and B, purple). Because the B+4 residue is not required for binding to PCNA (27), we postulated that Asp-122 and/or Glu-124 contact CRL4Cdt2 and help recruit it to the PCNA·PIP degron complex.
FIGURE 4.
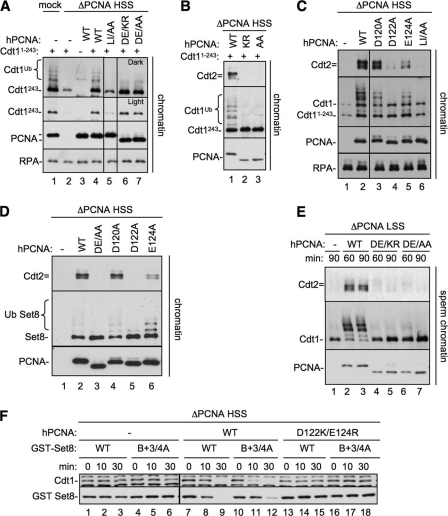
Identification of a PCNA residue that is essential for CRL4Cdt2-mediated destruction. A, an image of the PCNA-p21 co-crystal structure (24) was generated using PDB accession number 1AXC and PyMOL (available on-line). PIP box residues of p21 are shown in green, the B+4 of p21 is shown in blue, Leu-126 and Ile-128 (of the PCNA-79 mutant) are shown in red, and PCNA acidic residues Asp-122 and Glu-124 are shown in purple. B, sequence alignment of the interdomain connector loop of PCNA (amino acids 119–133) from different species. Leu-126 and Ile-128 are shown in red; Asp-122 and Glu-124 are shown in purple. C, PCNA-depleted HSS was supplemented with recombinant human PCNAWT, PCNADE/AA (PCNAD122A/E124A), PCNAD122A, PCNAD120A, or PCNAE124A. Recombinant Set8WT, Cdt11–243, and 5 ng/μl MMS plasmid were also added to the extract. Reactions were stopped at the indicated times and blotted for Cdt1 (both endogenous and Cdt11–243) or Set8. Samples were run on two separate gels that were processed under the same conditions. Additionally, PCNAWT samples were run on both gels, exposures were matched, and similar exposures were used.
We first tested whether Asp-122 and Glu-124, as well as another nearby acidic residue, Asp-120, are required for destruction of CRL4Cdt2 substrates. Asp-120, Asp-122, and Glu-124, or Asp-122 and Glu-124, were mutated to alanines, and the resulting proteins (PCNAD120A, PCNAD122A, PCNAE124A, and PCNADE/AA) were purified (supplemental Fig. S2A). PCNA-depleted HSS was supplemented with the different recombinant PCNA proteins, damaged DNA, as well as Set8 and Cdt11–243, an N-terminal fragment of Cdt1 that is destroyed by the same mechanism as Cdt1WT (27). Although PCNAD120A behaved essentially like PCNAWT, PCNAE124A displayed a noticeable, but minor defect, especially in Set8 destruction (Fig. 4C, lanes 17–20). Strikingly, PCNAD122A was completely inactive for destruction of Cdt1, Cdt11–243, and Set8 (Fig. 4C, lanes 9–12). PCNAD122A was also unable to support efficient Xic1 destruction (supplemental Fig. S2C). As expected, PCNADE/AA was unable to support CRL4Cdt2 activity in HSS extracts (Fig. 4C and supplemental Fig. S2B). In addition, PCNADE/AA did not support replication-dependent Cdt1 destruction in the context of sperm chromatin replication carried out in LSS extracts (supplemental Fig. S3A).
We examined the role of other PCNA residues near the IDCL. Alanine substitution of His-44, which contacts the 5 and 6 positions of bound PIP boxes, caused a slight defect in destruction of Cdt1, due to deficient substrate binding to PCNA, whereas S42A and S230A, which reside on either side of the IDCL, had no effect.3
It was important to rule out the possibility that the D122A and E124A mutations in PCNA inhibited CRL4Cdt2 activity due to indirect effects on DNA replication. As shown in supplemental Fig. S2D, PCNADE/AA supported normal levels of M13 DNA replication in HSS, a model for leading strand synthesis in Xenopus egg extracts, implying that this mutant is also loaded normally on chromatin. PCNADE/AA was also fully competent for sperm chromatin replication in LSS (supplemental Fig. S3B). Together, our results show that the D122A and E124A residues of PCNA play a specific role in potentiating CRL4Cdt2 function.
Asp-122 Is Required for CRL4Cdt2 Docking, but Not Substrate Binding to PCNA
To determine why the Asp-122 residue of PCNA is required for CRL4Cdt2 function, we first examined whether it mediates binding of substrates to PCNA. We took advantage of the fact that Cdt11–243 only binds to chromatin via PCNA in a PIP box-dependent manner (27). MMS-treated DNA coupled to magnetic beads was added to PCNA-depleted HSS supplemented with Cdt11–243 and either buffer, PCNAWT, PCNADE/AA, or PCNADE/KR, a charge-reversal mutant that also failed to support CRL4Cdt2 function and behaved identically to PCNADE/AA (supplemental Figs. S2B and S3B). We also included PCNALI/AA, a PCNA mutant previously studied in Saccharomyces cerevisiae (25), which carries mutations in residues Leu-126 and Ile-128 in the IDCL of PCNA and does not bind well to PIP box proteins (24–26, 43). Each of the recombinant PCNAs bound to chromatin at similar levels (Fig. 5A, PCNA), suggesting that none of the mutations affected replication factor C-mediated loading of PCNA onto DNA. Indeed, it was previously reported that even PCNALI/AA is efficiently loaded onto DNA by replication factor C (25). Importantly, equal amounts of Cdt11–243 bound to DNA in the presence of PCNAWT, PCNADE/AA, and PCNADE/KR (Fig. 5A, lanes 4, 6, and 7). In contrast, PCNAIL/AA did not support Cdt1 loading above background levels, as expected, based on previous studies using DNA polymerases (Fig. 5A, compare lanes 2 and 5) (25, 26). In conclusion, the defect in CRL4Cdt2 function observed with mutations in Asp-122 and Glu-124 was not due to a failure to recruit the substrate to chromatin-bound PCNA.
FIGURE 5.
PCNA Asp-122 is required for CRL4Cdt2 recruitment, but not substrate binding. A, Cdt11–243 binding to PCNA mutants. PCNA-depleted HSS was supplemented with 1-kb MMS DNA, methyl ubiquitin, Cdt11–243, and either buffer, PCNAWT, PCNALI/AA (PCNAL126A/I128A), PCNADE/AA (PCNAD122A/E124A), or PCNADE/KR (PCNAD122K/E124R). After 10 min, chromatin was recovered from extract, washed with 200 mm KCl-ELB, and blotted for the indicated proteins. All samples were run on the same gel, but some irrelevant lanes were removed. Note that PCNADE/AA and PCNADE/KR migrate faster than PCNAWT likely due to the change in charges. B, Cdt2 recruitment to chromatin by PCNA mutants. PCNA-depleted HSS was supplemented with 1 kb of immobilized MMS DNA, methyl ubiquitin, Cdt11–243, and either buffer, PCNAWT, PCNADE/AA (PCNAD122A/E124A), or PCNADE/KR (PCNAD122K/E124R). After 10 min chromatin was recovered from extract, washed, and blotted for the indicated proteins. C, PCNA-depleted HSS was supplemented with 1 kb of MMS DNA, methyl ubiquitin, Cdt11–243, and either buffer, PCNAWT, PCNAD120A, PCNAD122A, PCNAE124A, or PCNALI/AA. After 10 min chromatin was recovered from extract, washed, and blotted for the indicated proteins. All samples were run on the same gel, but some irrelevant lanes were removed. D, PCNA-depleted HSS was supplemented with 1 kb of MMS DNA, methyl ubiquitin, Set8WT, and either buffer, PCNAWT, PCNADE/AA, PCNADE/KR, PCNAD120A, PCNAD122A, PCNAE124A, or PCNALI/AA. After 10 min chromatin was recovered from extract, washed, and blotted for the indicated proteins. E, PCNA-depleted LSS was supplemented with sperm chromatin, methyl ubiquitin and buffer, PCNAWT, PCNADE/KR, or PCNADE/AA. After 60 or 90 min, chromatin was recovered from extract, washed, and blotted for the indicated proteins. F, PCNA-depleted HSS was supplemented with MMS plasmid, Set8WT, or Set8B+3/4A and either buffer, PCNAWT, or PCNADE/KR. The reactions were stopped at the indicated times and blotted for Cdt1 or Set8. Samples were run on two separate gels that were processed under the same conditions. Additionally, one sample set was run on both gels, exposures were matched, and similar exposures were used.
We next examined CRL4Cdt2 recruitment to chromatin in the presence of the PCNA mutants. MMS-treated 1-kb DNA conjugated to magnetic beads was added to PCNA-depleted extract supplemented with recombinant PCNAs and Cdt11–243. Unlike PCNAWT, PCNADE/KR and PCNADE/AA were inactive for Cdt2 recruitment and Cdt11–243 ubiquitylation (Fig. 5B). PCNAD120A was unaffected, whereas PCNAD122A behaved like PCNADE/AA with respect to Cdt1 binding, CRL4Cdt2 recruitment, and ubiquitylation (Fig. 5C, lanes 3 and 4). Consistent with its intermediate effects on Cdt1 destruction, PCNAE124A supported intermediate levels of Cdt2 recruitment (Fig. 5C, lane 5). Similar results were observed for Set8 (Fig. 5D). PCNADE/AA and PCNADE/KR also did not support Cdt2 recruitment or Cdt1 ubiquitylation in the context of sperm chromatin replication (Fig. 5E). Together, these data show that Asp-122 is essential and Glu-124 is important for substrate-dependent CRL4Cdt2 recruitment to chromatin, whereas these residues have no role in mediating the binding of PIP box proteins to PCNA.
PCNA Residues Asp-122 and Glu-124 Are Required for CRL4Cdt2-mediated Destruction Independently of the B+4 Residue
Based on the above results, we postulated that CRL4Cdt2 binds directly to residues in the PIP degron (B+4), as well as in PCNA (Asp-122 and Glu-124). However, one alternative explanation was that Asp-122 and Glu-124 do not directly contact CRL4Cdt2 (or an unknown ligase cofactor) but rather function to position B+4 for interaction with the ligase. To address this possibility, we examined whether mutations in Asp-122 and Glu-124 still affect destruction of a CRL4Cdt2 substrate lacking the B+4 residue. This experiment was not possible in the context of Cdt1, where mutation of B+4 alone completely eliminates destruction (27). Therefore, we examined the destruction of Set8B+3/4A in PCNA-depleted extracts supplemented with PCNAWT or PCNADE/KR. As shown in Fig. 5F, the intermediate level of Set8B+3/4A destruction was completely abolished in the presence of PCNADE/KR (compare lanes 10–12 with 16–18). Thus, PCNA residues 122 and 124 are critical for CRL4Cdt2-mediated destruction even in the absence of the B+4 residue, indicating that residues on PCNA are directly involved in ligase recruitment.
PCNA Residue Asp-122 Is Required for CRL4Cdt2-mediated Destruction in Fission Yeast
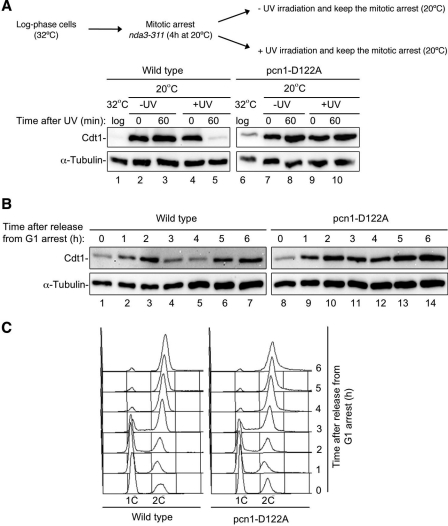
In Schizosaccharomyces pombe, CRL4Cdt2 targets Cdt1 for destruction in a PCNA-dependent manner during S phase and after DNA damage (39, 44). To determine whether the function of aspartic acid 122 in PCNA is conserved and whether it affects endogenous CRL4Cdt2 substrates in an in vivo model, we made the D122A mutation in the S. pombe PCNA gene, pcn1. Upon UV irradiation, cells expressing wild-type PCNA destroyed Cdt1, while cells expressing PCNAD122A did not (Fig. 6A, compare lanes 5 and 10). In addition, analysis of Cdt1 levels in cells released from a G1 arrest showed that the S phase destruction of Cdt1 is inhibited in the mutant (Fig. 6B, compare lanes 4 and 5 to lanes 11 and 12). Stabilization of TAP-tagged Cdt1 by PCNAD122A appeared to cause a minor delay in either S phase entry or progression through S phase (Fig. 6C, compare 3-h time points), but this could not explain the effect on destruction. Similar results for Cdt1 stabilization were seen in cells released from a mitotic block (supplemental Fig. S4). Therefore, as in Xenopus egg extracts, Asp-122 in S. pombe PCNA is required to support CRL4Cdt2 activity during replication and after DNA damage.
FIGURE 6.
The Asp-122 residue of PCNA is required for CRL4Cdt2-mediated destruction in fission yeast. A, Western blot analysis of Cdt1-TAP levels in cell extracts from a wild-type strain (2069) and a pcn1-D122A mutant (2640) prepared after UV irradiation (100 J/m2) or mock irradiated. Both strains contain the nda3–311 allele and were arrested in mitosis by incubation at 20 °C for 4 h, UV-irradiated, and kept at the restrictive temperature to maintain the mitotic arrest and prevent entry into S phase. B, Cdt1-TAP levels in wild-type (1540) and pcn1-D122A (2660) in G1-arrested cells, subsequently released into the cell cycle. C, flow cytometric analysis of DNA contents of cells shown in B.
DISCUSSION
In this study we explore the molecular mechanism by which CRL4Cdt2 recognizes its substrates in the context of PCNA. We provide evidence that recognition by CRL4Cdt2 requires amino acids not only in the substrate's PIP degron, but also in PCNA itself. The simplest interpretation is that CRL4Cdt2 makes direct contacts with both polypeptides during substrate recognition. This mechanism appears to be conserved from humans to fission yeast.
The activity of most ubiquitin ligases is regulated either by the assembly of ligase subunits, or post-translational modification of the ligase or substrate. To our knowledge, CRL4Cdt2 is the first example of a ubiquitin ligase whose activity is regulated by the creation of a bipartite surface when a substrate interacts with another polypeptide. Specifically, the event that triggers destruction of CRL4Cdt2 substrates is the creation of a composite surface composed of PCNA and the substrate. Conversely, CRL1Tir-auxin (SCFTir1-auxin) (45) and CRL1Skp2-Cks1 (SCFSkp2-Cks1) (46) require ligase cofactor interactions to recognize their substrates. In the case of CRL1Tir1-auxin, binding of auxin to the substrate receptor Tir1 creates a surface on the ligase that mediates binding to and ubiquitylation of its substrates (45). Although ubiquitylation of the CRL1Skp2 substrate p27 requires prior formation of a complex between the substrate receptor Skp2 and its cofactor Cks1, this interaction is not the initiating event to trigger p27 destruction (46, 47). Rather, CRL1Skp2-Cks1-mediated proteolysis is promoted by a phosphorylation event on threonine 187 of the substrate p27, which mediates the p27-CRL1Skp2-Cks1 interaction (3, 46, 49).
Mechanism of Substrate Recognition by CRL4Cdt2
Our data suggest the following model for the assembly of the ternary PCNADNA·PIP degron·CRL4Cdt2 complex. First, substrates bind PCNADNA via their PIP degron, an event that does not require CRL4Cdt2 (27) and therefore almost certainly precedes binding of the ligase. Next, CRL4Cdt2 docks onto the PCNA·PIP degron complex, likely using the WD40-repeat-containing β-propeller of Cdt2. A model for the structure of CRL4Ddb2, which ubiquitylates xeroderma pigmentosum, complementation group C (XPC), in the context of nucleotide excision repair (50–52), provides a framework for the possible structure of CRL4Cdt2. Thus, like the β-propeller protein Ddb2, Cdt2 likely contacts the adaptor protein Ddb1 via a helix located near the bottom surface of its propeller (50, 53). Accordingly, the top surface of the propeller of Cdt2 would interact with the PCNA·PIP degron complex. We have shown that the B+4 residue within the PIP degron and at least one residue on PCNA that cradles B+4 are essential for stably recruiting CRL4Cdt2. It is presently unclear whether residues in the PIP degron other than B+4 or residues in PCNA other than Asp-122 and Glu-124 make contact with CRL4Cdt2. Our data suggest that Cdt2 contains a surface with an appropriate arrangement of positive and negative charges that binds the PCNA·PIP degron complex (Fig. 7). Importantly, because substrate recognition by CRL4Cdt2 has not been reconstituted with purified components, we cannot rule out the possibility that the binding of CRL4Cdt2 to the PCNA·PIP degron complex is indirect. However, given the direct binding of several other β-propeller WD40 proteins to substrate (50, 54–58), this appears unlikely.
FIGURE 7.
Putative model of CRL4Cdt2 recruitment to the PCNA·substrate complex. Substrate-dependent recruitment of CRL4Cdt2 to PCNADNA requires residues in the PIP degron (B+4), as well as PCNA Asp-122. PCNA Glu-124 also contributes to CRL4Cdt2 recruitment but is not essential. We speculate charged residues in Cdt2 make direct contacts with B+4, Asp-122, and Glu-124.
An important question is whether PCNA functions primarily as a match-maker that promotes interactions between CRL4Cdt2 and its substrates (as illustrated in Fig. 7), or whether it also regulates ubiquitin transfer allosterically, by inducing conformational changes in the substrate or ligase. Our identification of residues on PCNA that are specifically required to recruit CRL4Cdt2 to the PCNA·substrate complex provides strong evidence for the former view, although it leaves open the possibility that PCNA could play additional roles in ubiquitin transfer.
Recently, Yew and colleagues proposed a two-step recognition model in which Xic1 and CRL4Cdt2 bind independently to two different subunits of PCNA and only later come together for Xic1 ubiquitylation (Fig. 3A) (11). This conclusion was based in part on an experiment in which a hybrid Xic1 substrate was constructed that was thought to have well separated PCNA and Cdt2 recognition motifs (Fig. 3B). However, we show here that the motifs were in fact not well separated (Fig. 3, C and D). Together with the observation that short peptides derived from Cdt1 (59) and p21 (27) are sufficient to support CRL4Cdt2 recruitment and activity, the data strongly favor a model in which the PCNA- and CRL4Cdt2-binding activities in the substrate are closely linked.
Contribution of Positively Charged Residues Upstream of the PIP Box
In Cdt1, mutation of the B+4 residue completely abolished destruction (27, 28), whereas in Xic1 and Set8, destruction was slowed but not eliminated (this report). In all cases, the mutation dramatically reduced the recruitment of CRL4Cdt2 to the PCNA·PIP degron complex. Importantly, the residual destruction of Set8B+3/4A (Fig. 5F) and Xic1B+4A (data not shown) still required Asp-122, arguing that this residue does not merely function to position the B+4 residue for recognition by CRL4Cdt2. Notably, the PIP degron of Cdt1 is located at the extreme N terminus of the protein, whereas in Set8, Xic1, and p21, this is not the case. The latter class of substrates also contains a cluster of basic residues immediately upstream of the PIP box. Mutation of these residues alone didn't interfere with destruction of Xic1. However, when they were mutated in combination with B+4, Xic1 was no longer degraded. The upstream basic residues are likely to contribute to the free energy of ternary complex formation, but this contribution is only readily detectable when the PIP degron is otherwise compromised through mutation of B+4. The discovery that Set8 and Xic1 lacking the B+4 residue can be inefficiently destroyed at reduced rates suggests that CRL4Cdt2 could also modify proteins that lack the B+4. However, the absence of this residue would have to be compensated for by other features to enhance CRL4Cdt2 recruitment, as illustrated by the basic residues upstream of the Xic1 PIP box. In addition, we know that the +5 downstream basic residue near the B+4 enhances PCNA binding, which contributes to efficient CRL4Cdt2-mediated destruction (13, 24, 27, 28). In fact, if the dimensions of the Cdt2 β-propeller are similar to those of Ddb2, separate areas of the “top” surface of Cdt2 could simultaneously contact the B+4 and upstream basic residues in the p21 PIP degron·PCNA complex.
Recently two distinct E2 ubiquitin-conjugating enzymes were identified that mediate ubiquitylation of substrates with CRL4Cdt2 (60). UBCH8 cooperates with CRL4Cdt2 to target p21 and Set8, whereas UBE2G1 and UBE2G2 collaborate with CRL4Cdt2 to ubiquitylate Cdt1. During polyubiquitin chain formation, E2s can recognize the surface of ubiquitin or the substrate near the lysine to be ubiquitylated (48, 61–64). Therefore, given the different PIP box locations and additional contributions of upstream residues of CRL4Cdt2 substrates, it is tempting to speculate that these different recognition determinants could contribute to the use of distinct E2s for Cdt1 versus p21 and Set8.
New Perspectives on the Degron
Our work raises interesting questions about the nature of degrons and the regulation of proteolysis. Because substrate recognition by CRL4Cdt2 requires residues in the PIP degron and in PCNA, PCNA could be considered part of the degron. However, to conform to the field's implicit understanding of the term, we propose that “degron” be reserved for recognition elements within the protein that gets destroyed. Eukaryotic cells contain hundreds of distinct ubiquitin ligases, most of which are completely uncharacterized, and we speculate that some of these might be regulated similarly to CRL4Cdt2. Thus, we propose that, among the thousands of transient protein-protein interactions that form during cellular growth and metabolism, some create a composite recognition surface that attracts a specific E3 ubiquitin ligase. Degron recognition could also involve the binding of protein substrates to other macromolecules such as nucleic acids, sugars, or lipids. The utility of this strategy is that it couples proteolysis to the final outcome of signaling events, which is usually the assembly of macromolecular complexes.
Supplementary Material
Acknowledgment
We thank Benjamin Morris for making the PyMOL models.
This work was supported, in whole or in part, by National Institutes of Health Grants GM80676 (to J. C. W.), GM082014 (to C. G. H.), and GM089150 (to R. C. C.). This work was also supported by Cancer Research UK Grant C814/A9035 (to S. E. K.).

This article contains supplemental Figs. S1–S4 and Tables S1 and S2.
B. Morris, C. G. Havens, and J. C. Walter, data not shown.
- CRL1
- Cullin ring ligase 1
- CRL4
- Cullin ring ligase 4
- MMS
- methyl methane sulfonate
- CDK
- cyclin-dependent kinase
- PCNA
- proliferating cell nuclear antigen
- IDCL
- interdomain connector loop
- HSS
- high speed supernatant
- LSS
- low speed supernatant
- TAP
- tandem affinity purification.
REFERENCES
- 1. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 2. Nguyen H., Gitig D. M., Koff A. (1999) Cell-free degradation of p27(kip1), a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol. Cell. Biol. 19, 1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montagnoli A., Fiore F., Eytan E., Carrano A. C., Draetta G. F., Hershko A., Pagano M. (1999) Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 13, 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu H. (2007) Cdc20. A WD40 activator for a cell cycle degradation machine. Mol. Cell 27, 3–16 [DOI] [PubMed] [Google Scholar]
- 5. Abbas T., Dutta A. (2011) CRL4Cdt2. Master coordinator of cell cycle progression and genome stability. Cell Cycle 10, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Havens C. G., Walter J. C. (2011) Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25, 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006) A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 [DOI] [PubMed] [Google Scholar]
- 8. Higa L. A., Banks D., Wu M., Kobayashi R., Sun H., Zhang H. (2006) L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5, 1675–1680 [DOI] [PubMed] [Google Scholar]
- 9. Sansam C. L., Shepard J. L., Lai K., Ianari A., Danielian P. S., Amsterdam A., Hopkins N., Lees J. A. (2006) DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20, 3117–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim D. H., Budhavarapu V. N., Herrera C. R., Nam H. W., Kim Y. S., Yew P. R. (2010) The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol. Cell. Biol. 30, 4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim Y., Starostina N. G., Kipreos E. T. (2008) The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22, 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishitani H., Shiomi Y., Iida H., Michishita M., Takami T., Tsurimoto T. (2008) CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283, 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abbas T., Shibata E., Park J., Jha S., Karnani N., Dutta A. (2010) CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., Zou L. (2010) CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oda H., Hubner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., Reinberg D. (2010) Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., Julien E. (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 18. Jørgensen S., Eskildsen M., Fugger K., Hansen L., Larsen M. S., Kousholt A. N., Syljuåsen R. G., Trelle M. B., Jensen O. N., Helin K., Sørensen C. S. (2011) SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 192, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shibutani S. T., de la Cruz A. F., Tran V., Turbyfill W. J., 3rd, Reis T., Edgar B. A., Duronio R. J. (2008) Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zielke N., Kim K. J., Tran V., Shibutani S. T., Bravo M. J., Nagarajan S., van Straaten M., Woods B., von Dassow G., Rottig C., Lehner C. F., Grewal S. S., Duronio R. J., Edgar B. A. (2011) Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S. H., Michael W. M. (2008) Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol. Cell 32, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C., Poitelea M., Watson A., Yoshida S. H., Shimoda C., Holmberg C., Nielsen O., Carr A. M. (2005) Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 24, 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 24. Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 25. Eissenberg J. C., Ayyagari R., Gomes X. V., Burgers P. M. (1997) Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol. Cell. Biol. 17, 6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang P., Sun Y., Hsu H., Zhang L., Zhang Y., Lee M. Y. (1998) The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase δ. J. Biol. Chem. 273, 713–719 [DOI] [PubMed] [Google Scholar]
- 27. Havens C. G., Walter J. C. (2009) Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michishita M., Morimoto A., Ishii T., Komori H., Shiomi Y., Higuchi Y., Nishitani H. (2011) Positively charged residues located downstream of PIP box, together with TD amino acids within PIP box, are important for CRL4(Cdt2)-mediated proteolysis. Genes Cells 16, 12–22 [DOI] [PubMed] [Google Scholar]
- 29. Lebofsky R., Takahashi T., Walter J. C. (2009) DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol. 521, 229–252 [DOI] [PubMed] [Google Scholar]
- 30. Walter J., Newport J. (2000) Initiation of eukaryotic DNA replication. Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5, 617–627 [DOI] [PubMed] [Google Scholar]
- 31. Walter J., Sun L., Newport J. (1998) Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1, 519–529 [DOI] [PubMed] [Google Scholar]
- 32. Arias E. E., Walter J. C. (2005) Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19, 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arias E. E., Walter J. C. (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 34. Kochaniak A. B., Habuchi S., Loparo J. J., Chang D. J., Cimprich K. A., Walter J. C., van Oijen A. M. (2009) Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 284, 17700–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stokes M. P., Michael W. M. (2003) DNA damage-induced replication arrest in Xenopus egg extracts. J. Cell Biol. 163, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gregan J., Lindner K., Brimage L., Franklin R., Namdar M., Hart E. A., Aves S. J., Kearsey S. E. (2003) Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell 14, 3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang X., Gregan J., Lindner K., Young H., Kearsey S. E. (2005) Nuclear distribution and chromatin association of DNA polymerase α-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast. BMC Mol. Biol. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindner K., Gregán J., Montgomery S., Kearsey S. E. (2002) Essential role of MCM proteins in premeiotic DNA replication. Mol. Biol. Cell 13, 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ralph E., Boye E., Kearsey S. E. (2006) DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 7, 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. You Z., Harvey K., Kong L., Newport J. (2002) Xic1 degradation in Xenopus egg extracts is coupled to initiation of DNA replication. Genes Dev. 16, 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chuang L. C., Yew P. R. (2005) Proliferating cell nuclear antigen recruits cyclin-dependent kinase inhibitor Xic1 to DNA and couples its proteolysis to DNA polymerase switching. J. Biol. Chem. 280, 35299–35309 [DOI] [PubMed] [Google Scholar]
- 42. Chuang L. C., Zhu X. N., Herrera C. R., Tseng H. M., Pfleger C. M., Block K., Yew P. R. (2005) The C-terminal domain of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1, is both necessary and sufficient for phosphorylation-independent proteolysis. J. Biol. Chem. 280, 35290–35298 [DOI] [PubMed] [Google Scholar]
- 43. Gomes X. V., Burgers P. M. (2000) Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19, 3811–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guarino E., Shepherd M. E., Salguero I., Hua H., Deegan R. S., Kearsey S. E. (2011) Cdt1 proteolysis is promoted by dual PIP degrons and is modulated by PCNA ubiquitylation. Nucleic Acids Res. 39, 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 46. Hao B., Zheng N., Schulman B. A., Wu G., Miller J. J., Pagano M., Pavletich N. P. (2005) Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol. Cell 20, 9–19 [DOI] [PubMed] [Google Scholar]
- 47. Xu S., Abbasian M., Patel P., Jensen-Pergakes K., Lombardo C. R., Cathers B. E., Xie W., Mercurio F., Pagano M., Giegel D., Cox S. (2007) Substrate recognition and ubiquitination of SCFSkp2/Cks1 ubiquitin-protein isopeptide ligase. J. Biol. Chem. 282, 15462–15470 [DOI] [PubMed] [Google Scholar]
- 48. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sitry D., Seeliger M. A., Ko T. K., Ganoth D., Breward S. E., Itzhaki L. S., Pagano M., Hershko A. (2002) Three different binding sites of Cks1 are required for p27-ubiquitin ligation. J. Biol. Chem. 277, 42233–42240 [DOI] [PubMed] [Google Scholar]
- 50. Scrima A., Konícková R., Czyzewski B. K., Kawasaki Y., Jeffrey P. D., Groisman R., Nakatani Y., Iwai S., Pavletich N. P., Thomä N. H. (2008) Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135, 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 52. Scrima A., Fischer E. S., Lingaraju G. M., Böhm K., Cavadini S., Thomä N. H. (2011) Detecting UV-lesions in the genome. The modular CRL4 ubiquitin ligase does it best! FEBS Lett. 585, 2818–2825 [DOI] [PubMed] [Google Scholar]
- 53. Li T., Chen X., Garbutt K. C., Zhou P., Zheng N. (2006) Structure of DDB1 in complex with a paramyxovirus V protein. Viral hijack of a propeller cluster in ubiquitin ligase. Cell 124, 105–117 [DOI] [PubMed] [Google Scholar]
- 54. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex. Destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 55. Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007) Structure of a Fbw7-Skp1-cyclin E complex. Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 [DOI] [PubMed] [Google Scholar]
- 56. Couture J. F., Collazo E., Trievel R. C. (2006) Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 13, 698–703 [DOI] [PubMed] [Google Scholar]
- 57. Schuetz A., Allali-Hassani A., Martín F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A. N., Arrowsmith C. H., Min J. (2006) Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 25, 4245–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han Z., Guo L., Wang H., Shen Y., Deng X. W., Chai J. (2006) Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell 22, 137–144 [DOI] [PubMed] [Google Scholar]
- 59. Senga T., Sivaprasad U., Zhu W., Park J. H., Arias E. E., Walter J. C., Dutta A. (2006) PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281, 6246–6252 [DOI] [PubMed] [Google Scholar]
- 60. Shibata E., Abbas T., Huang X., Wohlschlegel J. A., Dutta A. (2011) Selective ubiquitylation of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin-conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Mol. Cell. Biol. 31, 3136–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ye Y., Rape M. (2009) Building ubiquitin chains. E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jin L., Williamson A., Banerjee S., Philipp I., Rape M. (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rodrigo-Brenni M. C., Foster S. A., Morgan D. O. (2010) Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moreno S., Klar A., Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.