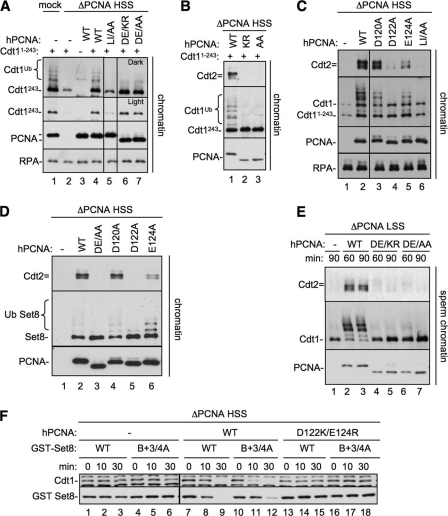
FIGURE 5.
PCNA Asp-122 is required for CRL4Cdt2 recruitment, but not substrate binding. A, Cdt11–243 binding to PCNA mutants. PCNA-depleted HSS was supplemented with 1-kb MMS DNA, methyl ubiquitin, Cdt11–243, and either buffer, PCNAWT, PCNALI/AA (PCNAL126A/I128A), PCNADE/AA (PCNAD122A/E124A), or PCNADE/KR (PCNAD122K/E124R). After 10 min, chromatin was recovered from extract, washed with 200 mm KCl-ELB, and blotted for the indicated proteins. All samples were run on the same gel, but some irrelevant lanes were removed. Note that PCNADE/AA and PCNADE/KR migrate faster than PCNAWT likely due to the change in charges. B, Cdt2 recruitment to chromatin by PCNA mutants. PCNA-depleted HSS was supplemented with 1 kb of immobilized MMS DNA, methyl ubiquitin, Cdt11–243, and either buffer, PCNAWT, PCNADE/AA (PCNAD122A/E124A), or PCNADE/KR (PCNAD122K/E124R). After 10 min chromatin was recovered from extract, washed, and blotted for the indicated proteins. C, PCNA-depleted HSS was supplemented with 1 kb of MMS DNA, methyl ubiquitin, Cdt11–243, and either buffer, PCNAWT, PCNAD120A, PCNAD122A, PCNAE124A, or PCNALI/AA. After 10 min chromatin was recovered from extract, washed, and blotted for the indicated proteins. All samples were run on the same gel, but some irrelevant lanes were removed. D, PCNA-depleted HSS was supplemented with 1 kb of MMS DNA, methyl ubiquitin, Set8WT, and either buffer, PCNAWT, PCNADE/AA, PCNADE/KR, PCNAD120A, PCNAD122A, PCNAE124A, or PCNALI/AA. After 10 min chromatin was recovered from extract, washed, and blotted for the indicated proteins. E, PCNA-depleted LSS was supplemented with sperm chromatin, methyl ubiquitin and buffer, PCNAWT, PCNADE/KR, or PCNADE/AA. After 60 or 90 min, chromatin was recovered from extract, washed, and blotted for the indicated proteins. F, PCNA-depleted HSS was supplemented with MMS plasmid, Set8WT, or Set8B+3/4A and either buffer, PCNAWT, or PCNADE/KR. The reactions were stopped at the indicated times and blotted for Cdt1 or Set8. Samples were run on two separate gels that were processed under the same conditions. Additionally, one sample set was run on both gels, exposures were matched, and similar exposures were used.