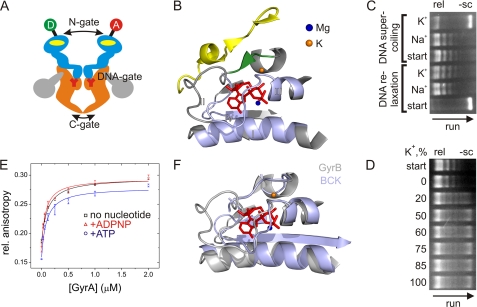
FIGURE 1.
DNA gyrase and influence of K+ and Na+ ions on topoisomerase activities and DNA binding. A, schematic representation of gyrase and the N-, DNA-, and C-gates. The active enzyme consists of two GyrA (orange and gray) and two GyrB (light blue) subunits. The yellow ovals indicate ATP-binding sites in GyrB, and Y represents the active site tyrosine in GyrA. The GyrA CTDs are depicted in gray. The circles indicate the attachment sites for donor (D) and acceptor (A) dyes at the N-gate. B, ATP-binding site in GyrB (Protein Data Bank code 1EI1, GyrB dimer in complex with ADPNP). The conserved GHKL motifs (I–IV) are depicted in light blue. The Mg2+ ion coordinating the phosphates is shown as a blue sphere, and the K+ ion is depicted in orange in the location where it is found in the protein kinase BCK (Protein Data Bank code 1GJV; see F). The K+ ion connects the α-helix contacting the phosphates of the bound nucleotide with the short β-strand (green) that forms a β-sheet with an α-strand from the second GyrB monomer (yellow). C, DNA relaxation (rel) and negative supercoiling (−sc) of DNA by gyrase in the presence of Na+ or K+. The arrow indicates the direction of the gel run. DNA relaxation (no ATP; start indicates negatively supercoiled DNA used as a substrate) is independent of K+, but negative DNA supercoiling (with ATP; start indicates relaxed DNA used as a substrate) requires K+ ions. D, dependence of negative DNA supercoiling on the K+ concentration. All reactions were performed at a constant ionic strength (100 mm, NaCl/KCl mixture) with 20 nm GyrA, 80 nm GyrB, 20 nm pUC18, and 1.5 mm ATP, varying the KCl concentration. The arrow indicates the direction of the gel run. The relaxed plasmid used as a substrate is shown in the start row. Sodium ions are not inhibitory for the supercoiling reaction. E, anisotropy titrations of an Alexa Fluor 488- and Alexa Fluor 546-labeled 60-bp DNA (7) in buffer containing Na+ with GyrA (8 μm GyrB added to form active gyrase). Kd values for the gyrase-DNA complex were 99 ± 12 nm (no nucleotide), 73 ± 16 nm (ATP), and 59 ± 11 nm (ADPNP). Error bars denote S.D. from three independent experiments. F, superposition of GyrB (gray; Protein Data Bank code 1EI1, GyrB dimer in complex with ADPNP) and BCK (light blue; Protein Data Bank code 1GJV), demonstrating the similar structures of the nucleotide-binding sites.