FIGURE 2.
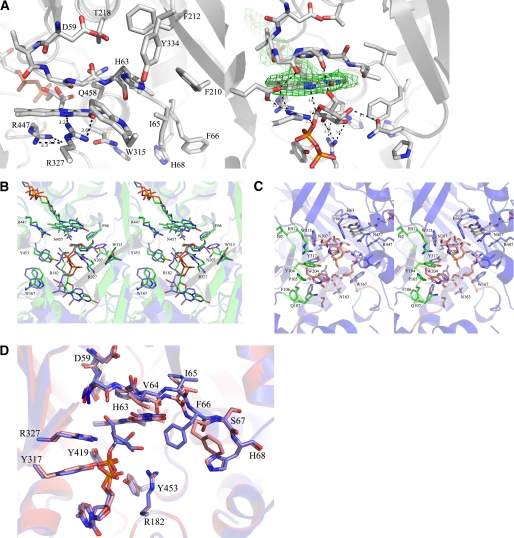
A, close-up of active sites from unliganded (left) and liganded AfUGM (right). Unliganded AfUGM, mobile loop III (residues 59–66) above the isoalloxazine ring with His-63 pointing into the highly conserved hydrophobic pocket. Arg-327 is at hydrogen bonding distance from N5 and O4 of FAD. Mobile loop III in the reduced AfUGM·UDP-Galp complex has flipped. His-63 sits above N5 of FADH2. Arg-327 is at hydrogen bonding distance of UDP-Galp. The Fo − Fc-electron density of the ligand (contoured at 3σ) is shown as a green wireframe in the right panel. B, stereodiagram comparison of the active sites from AfUGM·UDP-Galp (green) and DrUGM·UDP-Galp (blue). Labeling of active site residues is according to AfUGM sequence. C, stereodiagram of AfUGM active site with bound UDP-Galp. Residues shown in thick lines are within 4 Å from UDP-Galp. Residues in red are from closed mobile loops I and II. Residues in green (90–114) help to stabilize loop II and in positioning the uracil portion of UDP-Galp. D, overlay of reduced AfUGM·UDP-Galp (blue) on AfUGM·UDP (red). The binding mode of UDP is the same as the binding mode of the UDP moiety in the reduced UDP-Galp crystal structure. The only notable difference with the reduced UDP-Galp complex structure is that upon oxidation of the flavin the Phe-66 side chain moved outwards the active site and loop III at N3 side of the isoalloxazine ring moved ∼1 Å down.