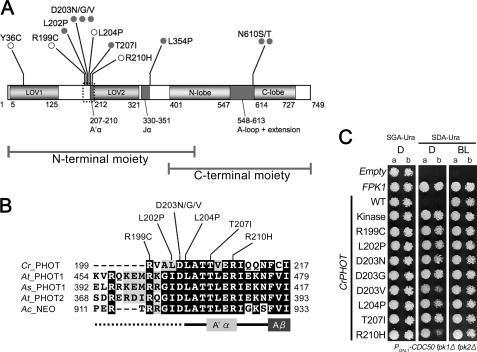
FIGURE 3.
CrPHOT mutants that restored yeast growth in darkness. A, schematic drawing of the domain structure of CrPHOT and locations of amino acid substitutions found in the present studies. Gray filled circles indicate mutations identified in clones with a single mutation. Open circles indicate mutations found in more than three independent clones with multiple mutations. B, sequence alignment of N-terminal flanking regions of LOV2 among different phototropin species. Cr_PHOT, C. reinhardtii PHOT; At_PHOT1, A. thaliana PHOT1; As_PHOT1, Avena sativa PHOT1; and At_PHOT2, A. thaliana PHOT2; Ac_NEO, Adiantum capillus-veneris NEOCHROME1. Sequences were aligned using the ClustalW and BOXSHADE program. C, growth of the PGAL1-CDC50 fpk1Δ fpk2Δ mutant expressing CrPHOT with single amino acid substitutions. Cells were treated as for Fig. 1B. Two concentrations of yeast cells (a, A600 = 0.004; b, A600 = 0.0008) were spotted.