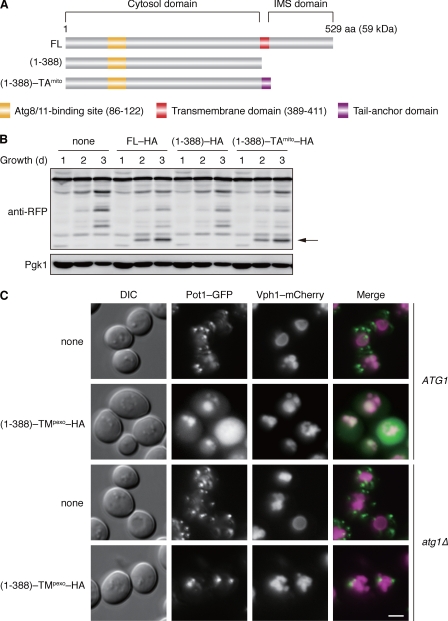
FIGURE 1.
Atg32 contains a domain compatible for degradation of peroxisomes. A, schematic representation of the domain structures of Atg32 (FL) and its truncated variants. (1–388) lacks the transmembrane and IMS domains, and (1–388) -TAmito consists of the cytosolic domain and a tail-anchor derived from Gem1, a mitochondrial outer membrane protein (39). B, cells expressing the wild-type or truncated variants of HA-tagged Atg32 (FL-HA, (1–388)–HA, or (1–388)-TAmito-HA) were grown in glycerol medium, collected at the indicated time points, and subjected to Western blotting. All strains are atg32-null derivatives expressing a mitochondrial matrix-localized DHFR-mCherry. The arrow depicts free mCherry generated by mitophagy. Pgk1 was monitored as a loading control. C, cells containing or lacking Atg1 (ATG1 or atg1Δ) were transformed with a low-copy, empty plasmid (none) or the one that encodes (1–388)-TMpexo-HA, an Atg32 cytosol domain anchored to the peroxisome via a TM domain derived from Pex15, a peroxisomal membrane protein (40). (1–388)-TMpexo-HA was expressed under the ATG32 promoter. All strains are atg32-null derivatives expressing both Pot1-GFP (peroxisomal marker) and Vph1-mCherry (vacuolar marker). Cells were grown in glycerol medium for 48 h and analyzed by fluorescence microscopy. Scale bar = 2 μm.