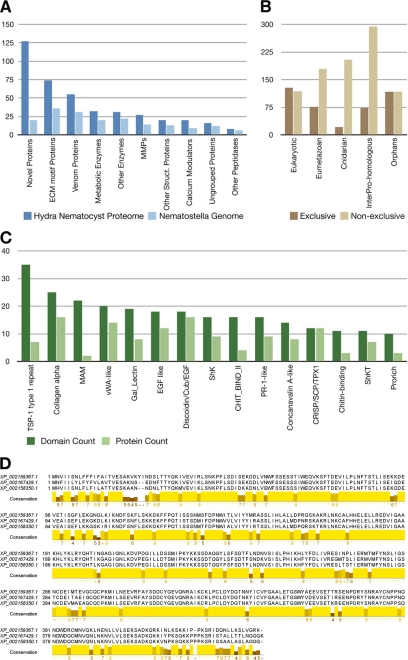
FIGURE 3.
A, homologs of Hydra nematocyst proteins identified in the Nematostella genome. MMPs, matrix metalloproteases. B, evolutionary distribution of the 410 nematocyst proteins. Nematocyst proteins have been assigned to categories based on the presence of significant BLAST hits and Pfam domains. The “exclusive” distribution refers to a hierarchical classification in which each protein was assigned to only one category. Proteins with significant BLAST hits in basal eukaryotes (T. adhaerens or M. brevicollis) have been classified as “eukaryotic.” The remaining proteins with significant similarity in H. sapiens, D. melanogaster, Danio rerio, C. elegans, or B. floridae were binned as “eumetazoan.” Proteins that were not classified as either eukaryotic or eumetazoan but with significant hits in the N. vectensis genome were classified as “cnidarian.” The remaining proteins were further classified as “InterPro-homologous” if they had a significant hit against the InterPro Database or as “orphans” otherwise. For the “non-exclusive” distribution, each nematocyst protein was assigned to every category where a significant hit could be identified. C, domain distribution of Hydra nematocyst proteins with a significant Pfam hit but no significant BLAST hit to eukaryotic, eumetazoan, or other cnidarian proteomes. Dark green, total number of occurrences of a domain in the data set; light green, number of proteins in which a domain occurs. vWA, von Willebrand factor A; SCP, sperm coat protein. D, alignment of the proteins corresponding to the second largest cluster of orphan proteins (see “Experimental Procedures”). The conserved region is predicted to be globular and thus likely to represent a previously uncharacterized protein domain. MAM, meprin, A-5 protein, receptor protein tyrosine phosphatase mu; Cub, complement C1r/C1s, Uegf, bone morphogenetic protein; ShkT, Stichodactyla toxin domain; Shk, Stichodactyla-like domain.