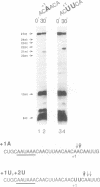
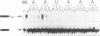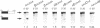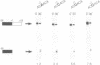Abstract
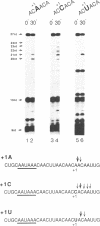
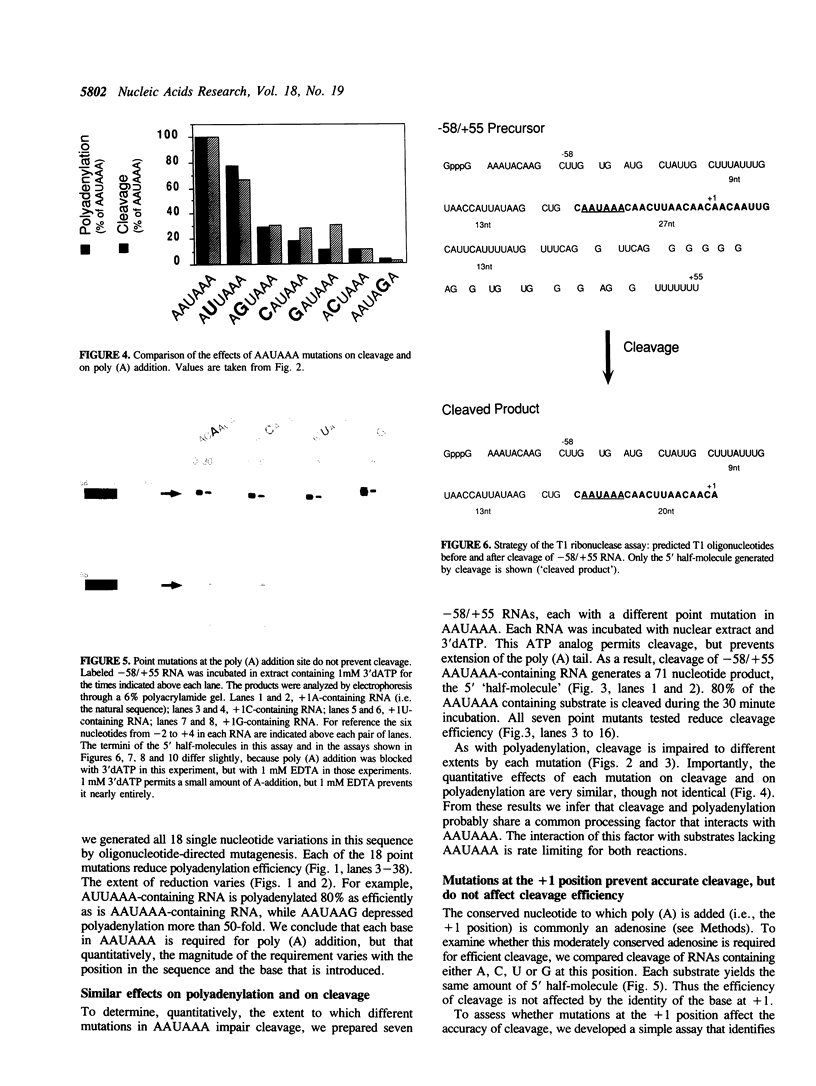
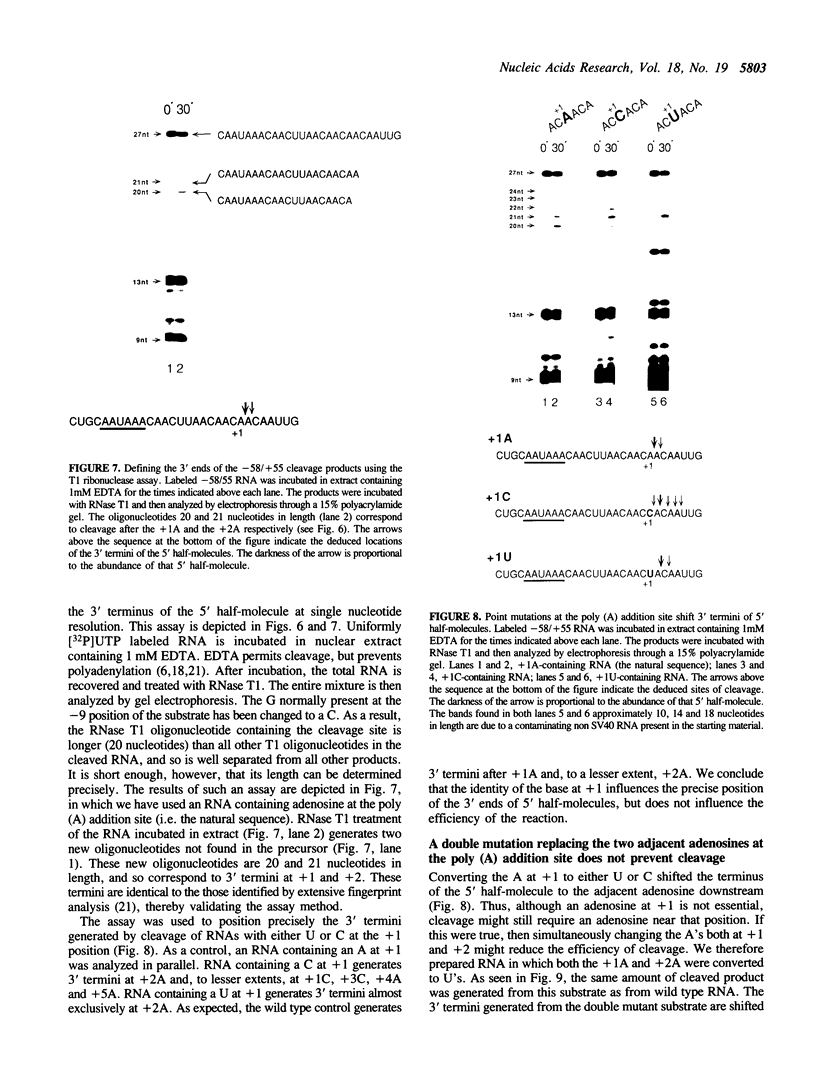
Three sequences in the vicinity of poly (A) addition sites are conserved among vertebrate mRNAs. We analyze the effects of single base changes in each position of AAUAAA and in the nucleotide to which poly (A) is added on 3' end formation in vitro. All 18 possible single base changes of the AAUAAA sequence greatly reduce addition of poly (A) to RNAs that end at the poly (A) addition site, and prevent cleavage of RNAs that extend beyond. The magnitude of reduction varies greatly with the position changed and the base introduced. For any given mutation, cleavage and polyadenylation are reduced to similar extents, strongly suggesting that the same factor interacts with AAUAAA in both reactions. Mutations at and near the conserved adenosine to which poly (A) is added disturb the accuracy, but not the efficiency, of 3' end formation. For example, point mutations at the conserved adenosine shift the 3' end of the most abundant 5' half-molecule downstream by a single nucleotide. The mechanism by which these mutations might exert their effects on the precision of 3' end formation are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G. M., McDevitt M. A., Nevins J. R. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988 May;2(5):578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- Gilmartin G. M., Nevins J. R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989 Dec;3(12B):2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J., Kimura S., Song B. J., Pastewka J., Gelboin H. V., Hardwick J. P. Sequence of two related P-450 mRNAs transcriptionally increased during rat development. An R.dre.1 sequence occupies the complete 3' untranslated region of a liver mRNA. J Biol Chem. 1986 Aug 15;261(23):10667–10672. [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Ali H., Nevins J. R. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol Cell Biol. 1985 Nov;5(11):2975–2983. doi: 10.1128/mcb.5.11.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Proudfoot N. J. A beginning to the biochemistry of polyadenylation. Trends Genet. 1988 Sep;4(9):243–245. doi: 10.1016/0168-9525(88)90028-5. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L. Accurate and specific polyadenylation of mRNA precursors in a soluble whole-cell lysate. Cell. 1983 Jun;33(2):595–605. doi: 10.1016/0092-8674(83)90440-3. [DOI] [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Yu H., Ryner L. RNA sequence containing hexanucleotide AAUAAA directs efficient mRNA polyadenylation in vitro. Mol Cell Biol. 1985 Feb;5(2):373–379. doi: 10.1128/mcb.5.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Skolnik-David H., Sharp P. A. Analysis of RNA cleavage at the adenovirus-2 L3 polyadenylation site. EMBO J. 1986 Aug;5(8):1929–1938. doi: 10.1002/j.1460-2075.1986.tb04446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Cheng T. C., Antonarakis S. E., Kazazian H. H., Jr Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985 Feb;4(2):453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sheets M. D., Stephenson P., Wickens M. P. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol Cell Biol. 1987 Apr;7(4):1518–1529. doi: 10.1128/mcb.7.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets M. D., Wickens M. Two phases in the addition of a poly(A) tail. Genes Dev. 1989 Sep;3(9):1401–1412. doi: 10.1101/gad.3.9.1401. [DOI] [PubMed] [Google Scholar]
- Skolnik-David H., Moore C. L., Sharp P. A. Electrophoretic separation of polyadenylation-specific complexes. Genes Dev. 1987 Sep;1(7):672–682. doi: 10.1101/gad.1.7.672. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Ryner L. C., Manley J. L. Four factors are required for 3'-end cleavage of pre-mRNAs. Genes Dev. 1989 Nov;3(11):1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Ryner L. C., Manley J. L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988 Mar 11;52(5):731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Wigley P. L., Sheets M. D., Zarkower D. A., Whitmer M. E., Wickens M. Polyadenylation of mRNA: minimal substrates and a requirement for the 2' hydroxyl of the U in AAUAAA. Mol Cell Biol. 1990 Apr;10(4):1705–1713. doi: 10.1128/mcb.10.4.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J., Pettine S. M., Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989 May 25;17(10):3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Formation of mRNA 3' termini: stability and dissociation of a complex involving the AAUAAA sequence. EMBO J. 1987 Jan;6(1):177–186. doi: 10.1002/j.1460-2075.1987.tb04736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]