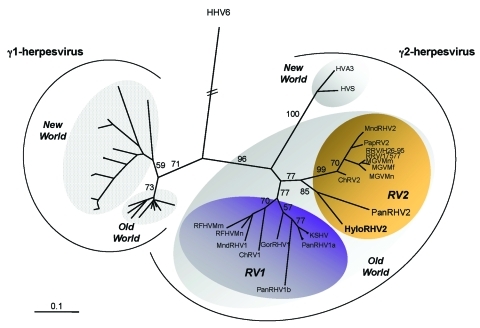
Figure 1.
Neighbor-joining protein distance tree for the best 130 amino acids of DNA polymerase residues alignment. DNA sequences (primers DFASA and GDTD1B) (8) were first translated, then aligned by using ClustalX (nonphylogeneticaly informative gaps were manually removed) and analyzed by using the PROTDIST and NEIGHBOR programs in PHYLIP (available from: http://evolution.genetics.washington.edu/phylip.html). Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 amino acid replacements per site. Numbers in each internal branch indicate the percentage of bootstrap samples (of 1,000) in which the cluster is supported (SEQBOOT). Previously published sequences included and their accession numbers are as follows: HyloRHV2 (AY465375); PanRHV2 (AF346490); KSHV (U75698, U93872 and AF005477); PanRHV1a (AF250879 and AF250880); PanRHV1b (AF250881 and AF250882); GorRHV1 (AF250886); MndRHV1 (AF282943); MndRHV2 (AF282937 to AF282940); herpesvirus saimiri (HVS) (M31122); ateline herpesvirus 3 (HVA3) (AF083424); Chlorocebus rhadinovirus 1 (ChRV1) (AJ251573); ChRV2 (AJ251574); retroperitoneal fibromatosis–associated herpesvirus strains from Macaca mulatta (AF005479) and M. nemestrina (AF005478) called here RFHVMm and RFHVMn, respectively; M. mulatta rhadinovirus RRV/17577 (AF083501); rhesus monkey rhadinovirus (RRV/H26-95) (AF029302); baboon γ-herpesvirus (PapRV2) (AY270026); Macaca γ-virus strains from M. mulatta (MGVMm) (AF159033), M. fascicularis (MGVMf) (AF159032), and M. nemestrina (AF159031); and HHV6A (X83413). Other sequences used to construct the phylogenetic trees (especially Epstein-Barr virus and related strains) have been published mainly by Ehlers et al. (9) and are named in the legend of Figure 2.