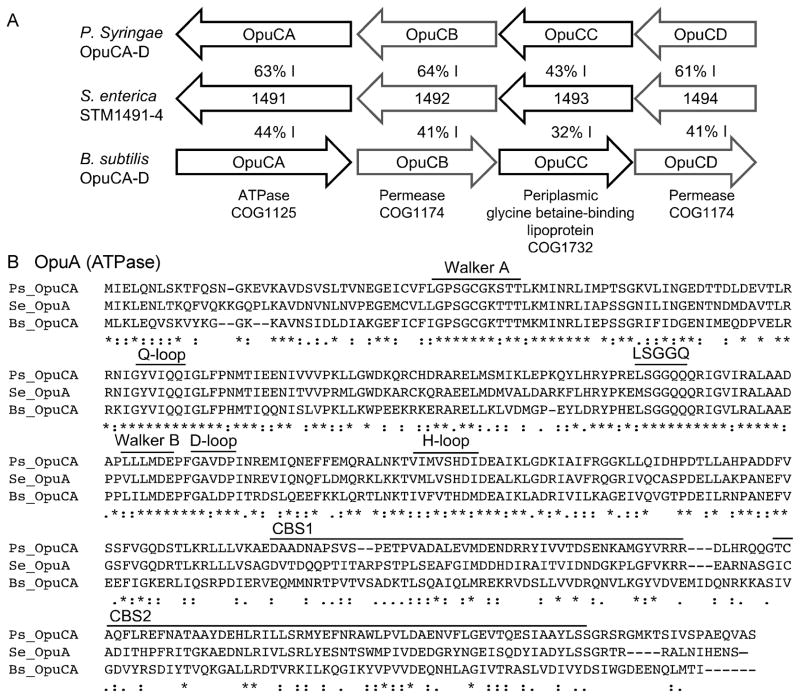
Figure 1. STM1491-4 is a putative glycine betaine importer.
A) The opu loci from Pseudomonas syringae (P.s. pathovar tomato strain DC3000), S. enterica STM1491-4 locus (S.e. serotype Typhimurium), and Bacillus subtilis (B.s. subspecies subtilis strain 168). Percent values between arrows indicates shared amino acid identity. Predicted encoded proteins and COG families are indicated at the bottom. B) Alignment of OpuA predicted ATPases from S. Typhimurium (SeT; STM1491, NP_460451.1), P. syringae (Ps; PSTPO_4575, AAO58021.1), and B. subtilis (Bs NP_391263.1). Conserved nucleotide-binding protein motifs are labeled, including the Walker A (P-loop), Q-loop, LSGGQ motif, Walker B, D-loop, and H-loop. CLUSTAL W (1.81) multiple sequence alignment and consensus key: single, fully conserved residue (*); conservation of strong groups (:); conservation of weak groups (.).