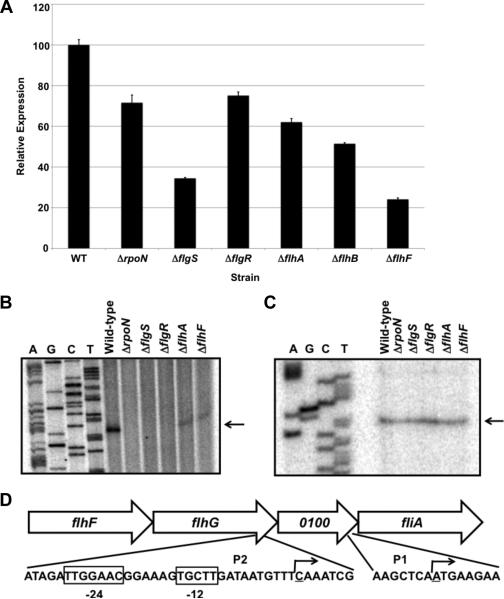
Figure 1. Analysis of fliA expression and transcriptional start sites in wild-type and mutant C. jejuni strains.
(A) Arylsulfatase assays measuring expression of a fliA::astA transcriptional fusion in wild-type C. jejuni 81-176 SmR and isogenic mutant strains lacking a component of the σ54 regulatory pathway. The amount of fliA::astA expression in each strain is relative to wild-type C. jejuni, which was set to 100 units. Error bars indicate standard error of the average arylsulfatase activity analyzed from three samples. The reporter activity in each mutant was significantly different from the activity in the wild-type strain (P-value < 0.05). (B and C) Primer extension analyses to identify transcriptional start sites for fliA. Two different primers were used to identify transcriptional start sites dependent (B) and independent (C) of the σ54 regulatory pathway. Primer extension reactions were performed with RNA from C. jejuni 81-176 SmR or isogenic mutant strains lacking a component of the σ54 regulatory pathway. Reactions were run alongside and to the right of a sequencing ladder generated with the same primer used in primer extension reactions. Arrows indicate transcriptional start sites. (D) Location of the transcriptional start sites for fliA. The transcriptional start site generated from the σ54-dependent promoter (P2) is located within the 3’ end of flhG. Boxed nucleotides indicate conserved -24 and -12 binding sites for σ54. The transcriptional start site from the σ54-independent promoter (P1) is located immediately upstream of fliA and within Cjj81176_0100. The underlined nucleotides and bent arrows indicate transcriptional start sites.