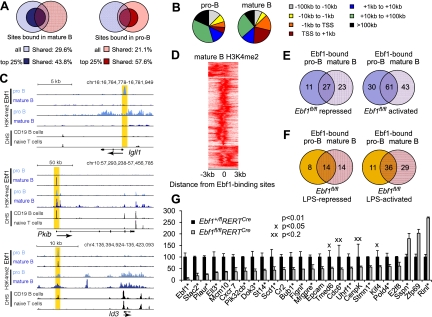
Figure 5.
Analysis of Ebf1 binding and Ebf1-mediated gene expression in mature B cells. (A) Overlap of genes occupied by Ebf1 in splenic B cells, as determined by ChIP-seq analysis, with Ebf1-bound genes in pro-B cells (Treiber et al. 2010). Shades of blue and red represent genes bound in pro-B cells and mature B cells, respectively. Dark colors represent Ebf1 targets in the top 25% of either library (see Supplemental Table S1). (B) Distribution of Ebf1-bound regions relative to annotated gene loci in mature B cells and pro-B cells. (C) Association of Ebf1-bound regions with H3K4me2 modification and DHS in Igll1, Pkib, and Id3 as representatives for genes bound by Ebf1 at different stages of differentiation. UCSC Genome Browser was used to visualize binding patterns. DHS data were obtained from ENCODE (http://genome.ucsc.edu/ENCODE). Vertical yellow band pattern indicates the position of Ebf1-binding sites. (D) Correlation of Ebf1 occupancy and H3K4me2 patterns in mature B cells. The heat map reflects the enrichment of H3K4 dimethylation in the vicinity of the top 25% Ebf1 peaks in mature B cells. The center of the Ebf1-binding sites in mature B cells is represented by 0. (E,F) Overlap of Ebf1-bound genes in pro-B cells and mature B cells (<100 kb from the TSS) that were also identified as functional targets by loss-of-function microarray analyses in unstimulated B cells (E) and LPS-stimulated B cells (F). The numbers represent Ebf1-bound genes that were identified as genes differentially expressed in Ebf1fl/flRERTCre versus Ebf1+/flRERTCre B cells. The identity of the genes and positions of the binding sites are shown in Supplemental Tables S5–S8. (G) Quantitative RT–PCR analysis of Ebf1-regulated targets that were identified in the microarray analysis. A representative set of 26 genes of the top 200 Ebf1-regulated target genes is shown, and fold expression values are relative to heterozygote control cells. Ebf1-bound genes among these targets are marked by an asterisk (see also Supplemental Fig. S3; Supplemental Tables S5, S6).