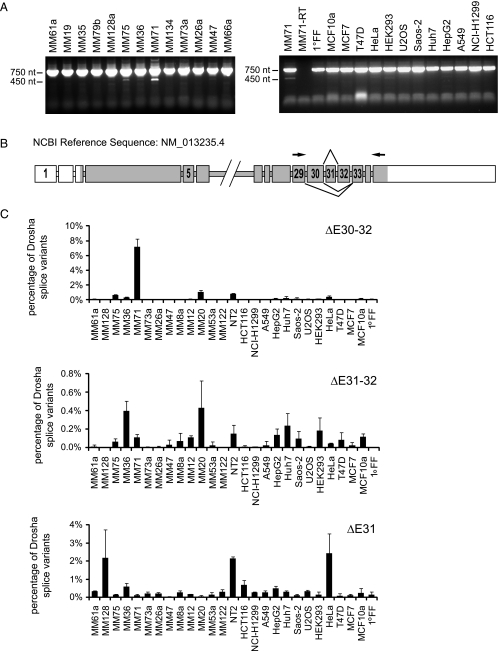
Figure 1.
Identification of alternative Drosha splice variants in melanoma cells. (A) Identification of alternative Drosha splice variants by reverse transcription. Drosha cDNA was amplified from exon 29 to 35 by reverse transcription in several cancer cell lines (MM = Ma-Mel). PCR products were visualized on a 1.2% agarose gel using ethidum bromide. The upper band is derived from wild-type Drosha. The lower bands are derived from splice variants lacking exons in the amplified region. (B) Schematic representation of the Drosha gene and derived alternative spliced mRNAs. Exon skipping changes the reading frame resulting in premature stop codons. Protein coding exons are shown in dark gray, noncoding exons in white. Arrowheads indicate positions of primers used to amplify exon 29 to 35. Introns are not drawn to scale. (C) Expression levels of splice variants lacking exons 31, 31 and 32, and 30 to 32 in different cancer cell lines quantified by quantitative RT-PCR in relation to total Drosha levels. Total Drosha levels were obtained by totaling the 2-ΔCt values normalized to RPLP0. The sum was set as 100%. One representative data set is shown; error bars, SEM.