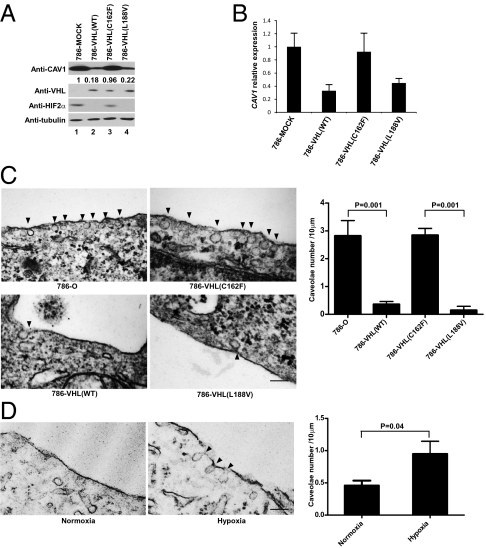
Fig. 3.
HIF promotes the formation of caveolae via positive regulation of CAV1. (A) Equal amounts of total cell lysates generated from the indicated isogenically matched subclones were resolved by SDS/PAGE and immunoblotted with the indicated antibodies. CAV1 signals were normalized to tubulin as measured by densitometry, and the CAV1 signal in 786-MOCK lysate (lane 1) was set arbitrarily at 1. (B) mRNA was extracted from the indicated cells, and the expression level of CAV1 mRNA was normalized against U1AsnRNP1 as measured by real-time PCR. The CAV1 level in 786-MOCK was set arbitrarily at 1. Error bars indicate SD. (C) (Left) The indicated cells were fixed and visualized by Hitachi H-7000 transmission electron microscopy. (Magnification: 100,000×.) Arrowheads indicate caveolae. (Scale bar.100 nm.) (Right) The number of caveolae structures on the cell membrane was quantified by transmission electron microscopy as described in Materials and Methods. Error bars indicate SD. P values were calculated using Student's t test. (D) (Left) 786-VHL cells maintained in normoxia or hypoxia for 48 h were fixed and visualized by Hitachi H-7000 transmission electron microscopy. (Magnification:100,000×.) Arrowheads indicate caveolae. (Scale bar: 100 nm.) (Right) The number of caveolae structures on the cell membrane was quantified by transmission electron microscopy as described in Materials and Methods. Error bars indicate SD. P values were calculated using Student's t test.