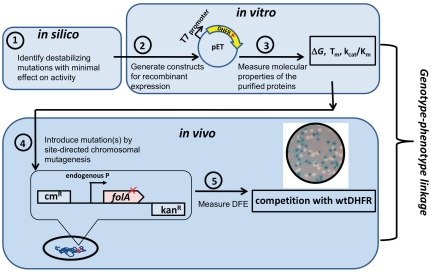
Fig. 1.
The experimental approach. (1) 10 DHFR residues, predominantly from the hydrophobic core and at least 4 Å from the NADPH and dihydrofolate binding sites, were chosen for mutagenesis based on the structural and phylogenetic predictions and published biophysical and biochemical data. (2) Sixteen single mutants (Table 1) were generated, cloned into pET vector, expressed, and purified (see SI Materials and Methods). (3) Gibbs free energy difference between folded and unfolded states (ΔG), apparent midtransition temperature of unfolding ( ), and catalytic parameters (kcat, Km) were measured (Table 1 and SI Materials and Methods). (4) A site-directed chromosomal mutagenesis method was developed to introduce in vitro characterized mutations into chromosomal folA gene of E. coli’s MG1655 strain without perturbing the gene’s regulatory region. In addition to 16 single mutant strains, 11 multiple mutant strains were generated by combining exhaustively four most destabilizing mutations (V75H, I91L, W133F, I155A) (Table S1). (5) Fitness effects of the introduced mutations (in total, 27 strains) were measured by growth competition of mutant strains with WT DHFR strain.
), and catalytic parameters (kcat, Km) were measured (Table 1 and SI Materials and Methods). (4) A site-directed chromosomal mutagenesis method was developed to introduce in vitro characterized mutations into chromosomal folA gene of E. coli’s MG1655 strain without perturbing the gene’s regulatory region. In addition to 16 single mutant strains, 11 multiple mutant strains were generated by combining exhaustively four most destabilizing mutations (V75H, I91L, W133F, I155A) (Table S1). (5) Fitness effects of the introduced mutations (in total, 27 strains) were measured by growth competition of mutant strains with WT DHFR strain.