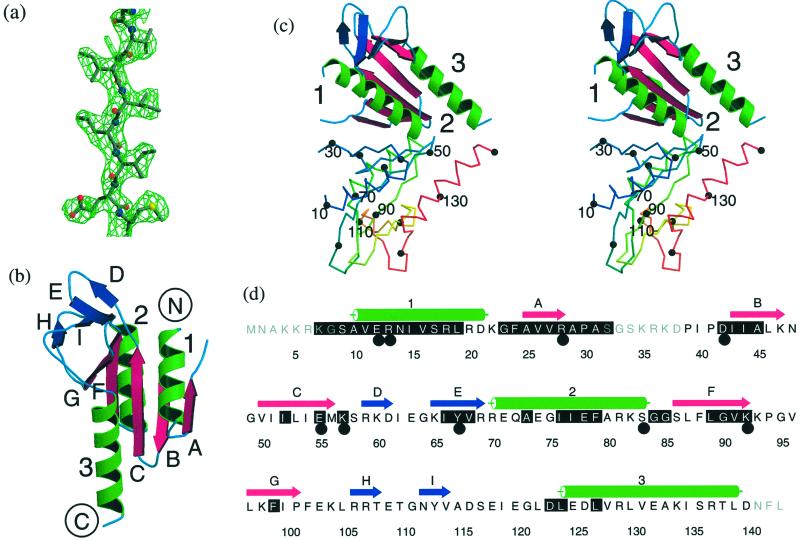
Figure 1.
The structure of S. solfataricus Hjc. (a) Solvent-flattened experimental electron density (1.3σ) for residues in strand C superimposed on the final model. (b) Ribbon representation of the Hjc monomer. The core β-sheet is shown as magenta arrows, α-helices are shown as green ribbon, and the peripheral β-sheet is shown as blue arrows. (c) Stereo representation of the Hjc dimer. One subunit is colored as above, with helices 1–3 labeled. The second subunit is a trace colored from blue at the N terminus to red at the C terminus. Every tenth Cα atom is marked by a black sphere and every twentieth is labeled. (d) The amino acid sequence of Hjc. Secondary structure elements are colored as above. Black shading marks residues that are highly conserved among Hjc family members. Large black circles mark residues for which mutation is deleterious to function. a–d were prepared by using MOLSCRIPT (36), RASTER3D (37), and ALSCRIPT (38). Secondary structure was assigned by using DSSP (39).