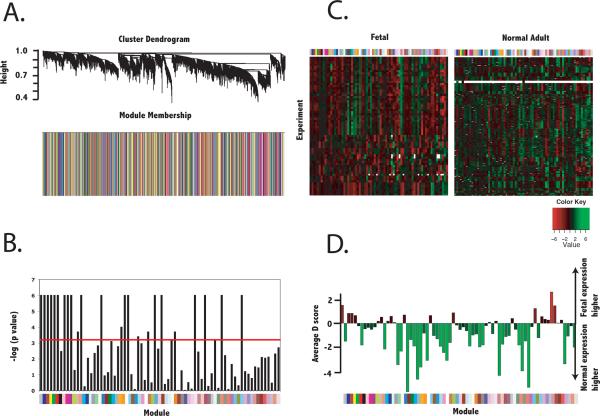
Figure 2.
Identification of gene coexpression modules specific to fetal tissue. Gene-gene adjacencies were first defined by the topological overlap, which quantifies the degree of shared network neighbors given by gene-wise Pearson correlation. Hierarchical clustering of adjacencies was first performed in the training dataset of 43 fetal samples and resulting coexpression modules were evaluated for significant reproducibility in the validation dataset of 21 fetal samples. The seventy-two valid fetal modules were evaluated for reproducibility in normal adult myocardium as well as differential expression. A.) Cluster dendrogram and barplot describing module membership (according to clusters derived in the training fetal dataset) for normal adult myocardium. B.) Reproducibility of valid fetal modules in normal adult myocardium. Colors below the bargraph correspond to the module membership. Red line indicates cutoff for statistical significance (p < 6.9× 10−4 or − log(p value) > 3.16). Modules below this cutoff were defined as having topology specific to fetal myocardium and defined as fetal modules. C.) Heatmap of average expression of each module in fetal and normal adult myocardium. Colors above the heatmaps correspond to module membership. D.) Average significance level of differential expression (average Significance Analysis of Microarrays d score) for each module. Negative values correspond to lower expression levels in normal than fetal myocardium.