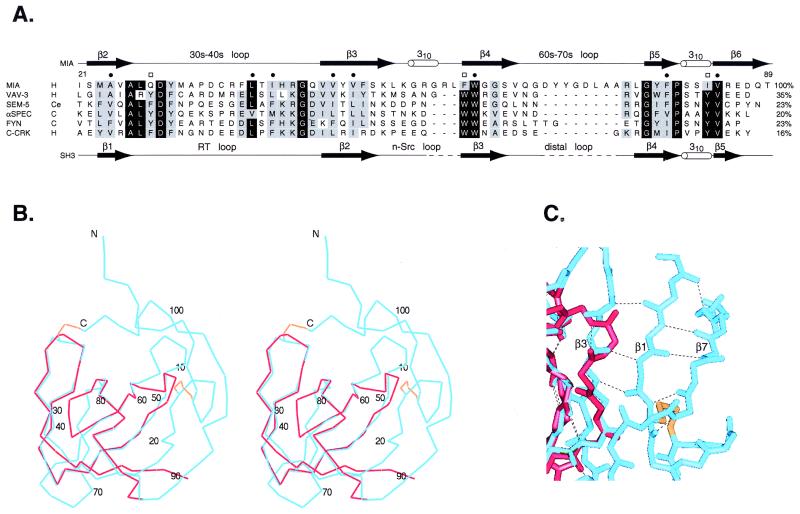
Figure 2.
Comparison of MIA to SH3 domains. (A) Sequence alignment of MIA with several SH3 domains. Species are abbreviated as follows: H, human; Ce, Caenorhabditas elegans; and C, chicken. Residues labeled with a bullet make up the conserved hydrophobic core in SH3 domains. Conserved SH3 residues labeled with a box make up a triad of aromatic residues arranged approximately linearly at the polyproline helix binding site. The secondary structure of MIA is shown above the alignment, and the secondary structure of the C-Crk SH3 domain is shown below the alignment. The percent identity relative to MIA is listed at the right of each sequence. (B and C) Comparison of the MIA structure with an SH3 domain. MIA is in blue, the Sem-5 SH3 domain is in red, and the MIA disulfide bonds are in gold. (B) Stereo view of the superposition of MIA with the Sem-5 SH3 domain. The rms deviation of the carbon-α positions is 0.61 Å, where 37 α-carbon pairs were used in the superposition. The numbering corresponds to human MIA. (C) View of the β3 strand extension in MIA relative to SH3 domains. Hydrogen bonding between β3 and β1 extends the length of strand β3.