Figure 3.
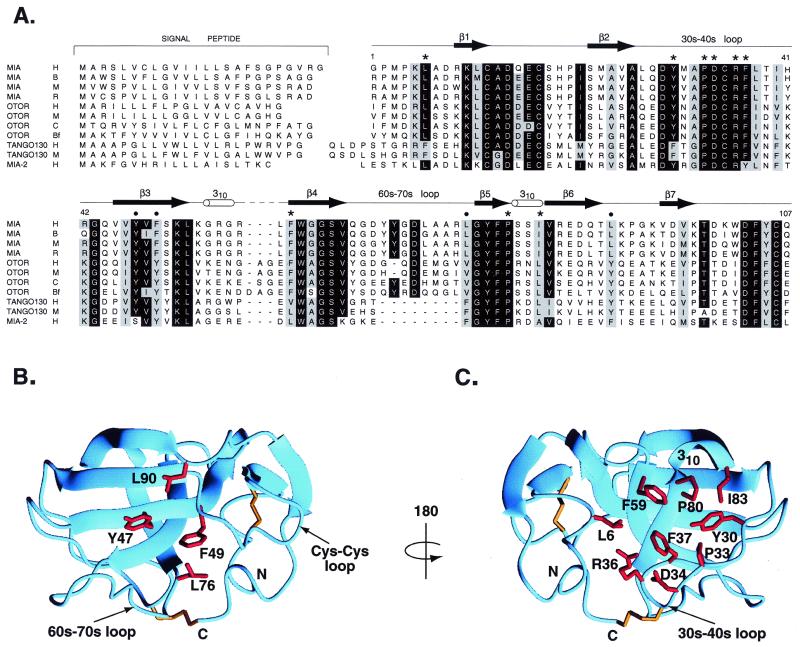
Sequence alignment of MIA with secreted homologues defines possible ligand binding sites. (A) Sequence alignment of MIA with secreted homologues. Species are abbreviated as follows: H, human; B, bovine; M, mouse; R, rat; C, chicken; and Bf, bullfrog. Numbering corresponds to human MIA, and the MIA secondary structure is shown. Bullets indicate a conserved patch located on one side of the molecule; these residues are displayed on the MIA structure in B. Stars indicate conserved surface residues on the opposite side of the molecule. These residues are shown on the MIA structure in C. In SH3 domains, the structurally analogous region to that shown in C is the polyproline helix binding site. Signal peptides were predicted by using the program signalp (34). MIA-2 is predicted by genscan from GenBank genomic sequence 7708219. tango 130 sequences are from U.S. patent WO 00/12762.