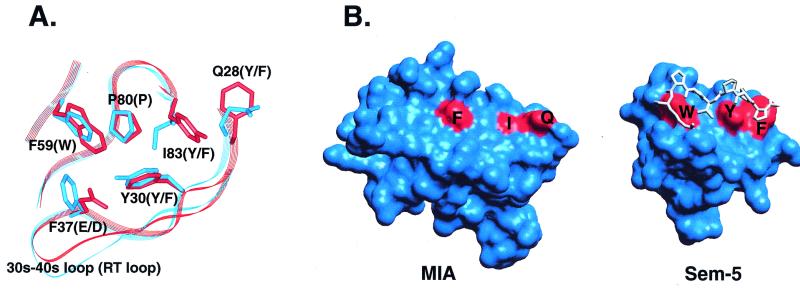
Figure 4.
Comparison of the SH3 polyproline helix binding site to the analogous region in MIA. (A) Superposition of the conserved residues in the polyproline helix binding site of Sem-5 (red) and the corresponding residues in MIA (blue). Four of six residues conserved in SH3 domains differ in MIA. Residue numbering corresponds to MIA. Residues listed in parentheses are those found in canonical SH3 domains. (B) View of the molecular surfaces of MIA (Left) and the Sem-5 SH3 domain (Right). The structure of Sem-5 is shown with its polyproline ligand bound. Sem-5 residues colored in red are those that make up a triad of conserved aromatic residues arranged approximately linearly on the molecular surface. The dissimilarity of the corresponding residues in MIA, F59, I83, and Q28 result in a much smoother molecular surface that most likely does not recognize polyproline helices.