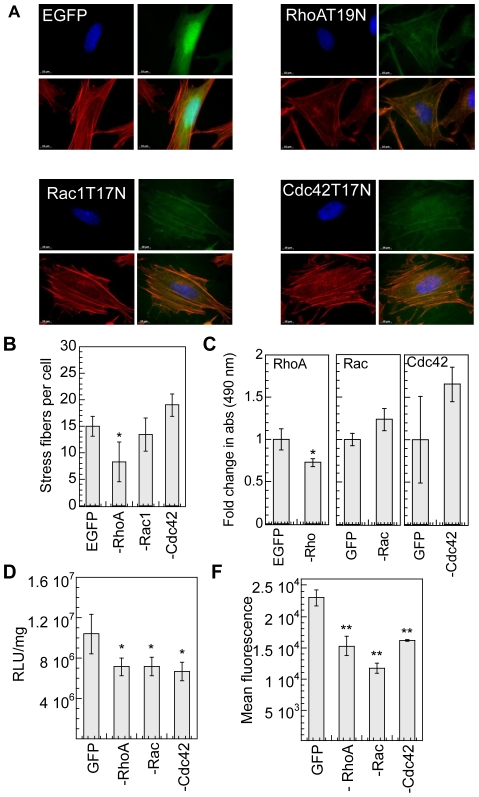
Figure 3. RhoGTPase inhibition using dominat negative gene products decreases transgene expression.
mMSCs were transiently transfected with dominant negative forms of RhoA, Rac1 or Cdc42 with a plasmid also containing EGFP. 24 hours post transfection, cells were replated on fibronectin coated plates and cultured for 16 hours prior to bolus transfection. Cells transfected with pEGFP were used as control. (A) Alexa488 conjugated phalloidin (green), and Hoechst dye (blue) staining. Images were taken with a Zeiss AxioObserver Z1 inverted microscope at 100× magnification. (B) Stress fiber quantification. Statistical analysis was done using a one-way Anova followed by the Dunnett Multiple Comparison test. The Anova p value was 0.0015. The symbol * represents a significant change in stress fibers with respect to untreated cells to the level of p<0.05. (C) Active Rho, Rac and Cdc42, was assessed for cells replated on fibronectin for 16 hours using GLISA assays specific for RhoA, Rac1,2,3 and Cdc42. Statistical analysis was done using the unpaired t-test (two tail p value). The symbol * represents a significant change to the level of p<0.05. (D) The transgene expression was analyzed 48 hours post transfection using luciferase assay and normalized with total protein analyzed using Peirce BCA assay. (E) Internalization was analyzed using YOYO-3 labeled polyplexes 2 hours post transfection using flow cytometry. A total of 10,000 events were analyzed per sample and the mean fluorescence of events positive for both GFP and YOYO-3 was analyzed. Statistical analysis for gene transfer and internalization was done using a one-way Anova followed by the Dunnett Multiple Comparison test. The Anova p values were 0.0219 and <0.0001 for transfection (D) and internalization (E) respectively. The symbols * and ** represents a significant change with respect to cells transfected with control plasmid (pEGFP) to the level of p<0.05 and p<0.01, respectively.