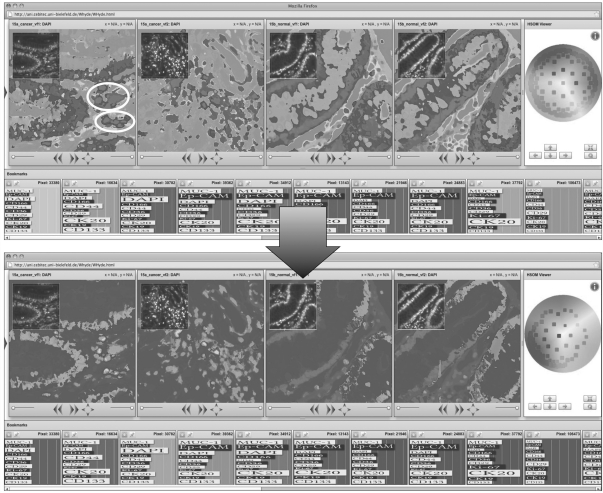
Fig. 3.
The H2SOM architecture provides the structural basis for a synchronized interactive dynamic pseudocoloring of TIS images. In the upper row, the four TIS images T(c1), T(c2), T(n1) and T(n2) from left to right. The bottom row shows a small set of selected bookmarked CIPRAs. On the right, the color disc is shown with its control buttons below. To change the coloring, the user can combine two functions. First, using the rotate-button, the user can turn the color disc so that of the H2SOM grid which is of less interest is mapped to the blue area (since human observers are more sensitive to non-blue colors). Second, the user can use the arrow buttons to change the Möbius projection, i.e. to move H2SOM nodes toward the center and squeeze the opposing nodes into a small cloud. In this example, the nodes from the upper right are moved to enhance the color contrast for a chosen region of interest in one image (marked with white ellipsoids).