Abstract
Active enkephalin and related peptide hormones or neurotransmitters are generated by proteolytic processing of inactive prohormone precursors. Little is known about the relative accessibilities of prohormone cleavage sites and conformations of subdomains that undergo proteolytic processing. Therefore, this study investigated the conformational features of the prohormone proenkephalin (PE) by rapid hydrogen-deuterium exchange mass spectrometry (DXMS). DXMS analyzes rates of hydrogen exchange of the polypeptide backbone of PE with deuterium from D2O (heavy water) by mass spectrometry, accomplished at sub-second and multisecond time periods. Results showed differential accessibilities of cleavage sites and adjacent subdomains of PE to the aqueous environment. Importantly, protease cleavage sites of PE with greater relative accessibilities correspond to sites most readily cleaved by processing proteases to generate active peptide neurotransmitters. For comparison, peptides derived from PE (by pepsin digestion) displayed greater accessibility to the solvent environment, illustrated by their higher rates of H-D exchange compared to that of intact PE protein. The more limited H-D exchange accessibilities of PE protein, compared to peptides derived from PE, indicate that PE possesses tertiary conformation. These results demonstrate that differential tertiary conformations of PE subdomains undergo ordered proteolytic processing to generate active enkephalin peptides for cell-cell communication in the nervous and endocrine systems.
Keywords: proenkephalin, prohormone, peptide neurotransmitter, peptide hormone, neuropeptide, hydrogen-deuterium exchange mass spectrometry, conformation, protease cleavage, aqueous environment
Introduction
Prohormone precursors must undergo proteolytic processing to generate small active peptide hormones and neurotransmitters, known as neuropeptides, that are essential for cell-cell signaling in the nervous and endocrine systems.1, 2 Proenkephalin (PE), like other prohormones, is processed at multiple cleavage sites to generate enkephalin peptides that participate in the control of analgesia,3–5 learning and memory,6, 7 and brain-regulated behaviors including drug addiction.8–10 PE contains 12 cleavage sites designated by dibasic residues in various heterocombinations and homocombinations of arginine and lysine, for example, Lys-Arg, Arg-Arg, and Lys-Lys that generate (Met)enkephalin (ME), (Leu)enkephalin (LE), ME-Arg-Gly-Leu, and ME-Arg-Phe.1 Proteolytic processing of PE and prohormones at dibasic processing sites is achieved by secretory vesicle processing endoproteases consisting of the cysteine protease cathepsin L1, 11, 12 and the subtilisin-like prohormone convertases (PC) PC2 and PC1/3, combined with the exopeptidases carboxypeptidase E and aminopeptidase B.1, 13, 14 The processed neuropeptides undergo regulated secretion for control of target cellular systems.
The multiple protease processing sites are located throughout PE, suggesting the tertiary conformation of PE may consist of cleavage site domains and other subdomains with differential accessibilities to the aqueous environment. Furthermore, processing of PE by sequential cleavages15, 16 also suggests the hypothesis that differential accessibilities of protease cleavage sites and intervening domains within PE occur. For these reasons, this study evaluated the hypothesized differential accessibilities of PE subdomains to the aqueous solvent environment by hydrogen-deuterium exchange mass spectrometry (DXMS), which measures rates of exchange of deuterium from heavy water (D2O) to the polypeptide backbone of the PE protein. Different rates of exchange among sub-regions in a polypeptide chain can indicate relative differences in their conformational features with respect to accessibilities and solvent access.17–24 For PE and other prohormones, it is hypothesized that unequal solvent accessibilities of PE subdomains participate in prohormone processing to generate active peptides.
The hypothesis that PE contains tertiary conformations predicts that its subdomains may have restricted accessibility to the solvent environment compared to short peptides derived from PE, which are predicted to possess greater accessibility to the aqueous environment than the same sequences within the intact PE. Therefore, this study also compared H-D exchange properties of intact PE with that of individual peptides derived from PE. Use of a quenched-flow system for measuring H-D exchange allowed determination of the dynamic and rapid nature of H-D exchange among PE domains, and even more rapid H-D exchange of peptides derived from PE.
H-D exchange results showed that PE displayed dynamic differences in relative accessibilities among its cleavage sites and intervening subdomains flanking the active enkephalin peptide sequences. Notably, the protease cleavage sites of PE that show greater relative accessibilities correspond to sites most readily cleaved by processing proteases to generate active peptide neurotransmitters.1 In contrast, peptides derived from the same PE (generated by pepsin) showed greater H-D exchange than intact PE. The more limited H-D exchange of intact PE indicates its tertiary conformational features. These H-D exchange properties illustrate that the tertiary conformational landscape of PE participates in its proteolytic processing to generate active enkephalin peptides for cell-cell communication.
Results
Rapid and differential H-D exchange rates of PE subdomains
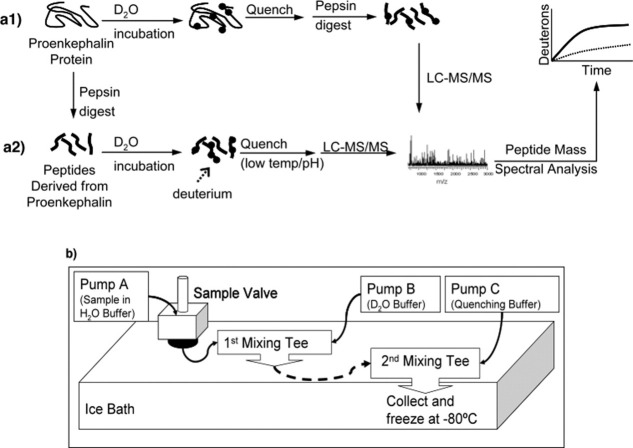
H-D exchange among subdomains of intact PE was achieved by incubation in heavy water (D2O), in time course studies, quenching, pepsin digestion, followed by peptide mass analyses by LC-MS/MS to determine incorporation of deuterium in pepsin-generated peptide fragments [Fig. 1(a1)]. For comparison, peptides derived from PE by pepsin digestion (rather than intact PE) were subjected to H-D exchange in time course studies [Fig. 1(a2)]. H-D exchange at rapid sub-second and multisecond time points required use of a quenched-flow system [Fig. 1(b)], which was necessary for the observation of more rapid H-D exchange properties of PE compared with other proteins that typically utilize longer H-D exchange times.17–24
Figure 1.
H-D exchange of PE and peptides derived from PE, with quenched flow system. a1 and a2: DXMS of intact PE and peptides derived from PE. DXMS (hydrogen-deuterium exchange mass spectrometer) was used to investigate H-D exchange properties of intact PE (part “a1”) and individual peptides derived from PE (by pepsin; part “a2”). In (a1), recombinant PE was subjected to incubation in D2O buffer in time course studies, followed by quenching, pepsin digestion, and LC-MS/MS for analyses of deuterons incorporated into PE regions. In (a2), the first step was to generate peptides derived from PE by pepsin digestion, and these peptides were then subjected to incubation in D2O buffer in time course studies followed by quenching and LC-MS/MS. (b) Quenched-flow system for rapid H-D exchange. A quenched-flow system was developed to obtain rapid sub-second and multisecond time points for H-D exchange data. Pump A delivers the sample in H2O buffer and pump B delivers the D2O buffer (buffer compositions are indicated in the methods) to a micro static mixing tee. Sub-second and multisecond H-D exchange periods were achieved by varying the tubing volume of the delay tube (dashed line in Fig. 2) that brings the mixture to a second mixing tee to which the quenching buffer is delivered by pump C. The time of exposure of the sample to D2O is determined by the volume of that particular tubing because the sample is in continuous flow at 0.4 mL/min prior to being quenched at the second mixing tee. These H-D exchange experiments were conducted in a water bath of 3.9°C at time points of 0.5, 1, 10, and 37.5 s.
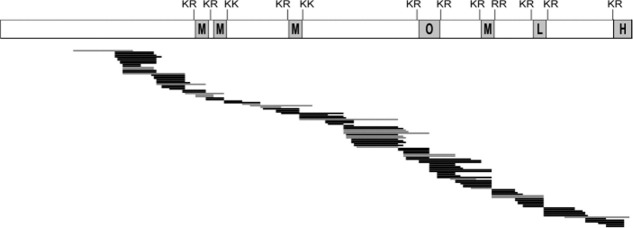
Pepsin digestion of PE and LC-MS/MS yielded excellent coverage of peptides identified (Fig. 2). Peptides included cleavage site domains of PE, and intervening domains between active enkephalin sequences. These pepsin digestion conditions were used for DXMS experiments in this study.
Figure 2.
Pepsin digestion of PE and mass spectrometry. The map of pepsin-generated peptides after H-D exchange illustrates the excellent coverage of peptides spanning the PE protein, including the active peptides within the PE precursor. The active peptides consist of (Met)enkephalin (M), M-Arg-Gly-Leu (O, octapeptide), (Leu)enkephalin (L), and M-Arg-Phe (H, heptapeptide). The pepsin-generated peptides shown in black lines were used for analyses of H-D exchange of intact PE and peptides derived from PE in Figures 3 and 4. The gray lines were observed in pepsin digests of PE, but were not used in deuteration maps of PE in Figures 3 and 4.
H-D exchange of intact PE reveals differential accessibilities of its subdomains
DXMS analyses of PE at the shortest time point of 0.5 s demonstrated H-D exchange occurring throughout the PE protein (Fig. 3). After incubation of PE in D2O, H-D exchange is illustrated by mapping pepsin-generated peptide fragments along the length of the PE polypeptide sequence. Time course experiments at 0.5, 1, 10, and 37 s of H-D exchange illustrated the increasing levels of deuteration of peptide domains of PE with longer incubation times (Fig. 3). Of interest was the observation that at the initial time points (0.5–10 s), differences in H-D exchange were observed throughout different subdomains of PE.
Figure 3.
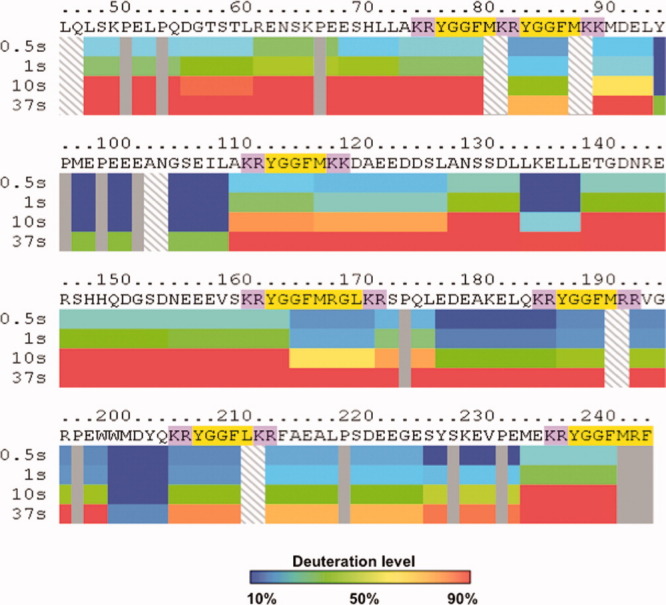
Differential H-D exchange among subdomains of intact PE indicated by DXMS time-course studies. Dynamic differences in the extent of deuteration are observed among different subdomains of PE. Time course studies incubated intact PE in D2O buffer at 0.5, 1, 10, and 37 s in DXMS experiments. Incorporation of deuterium from D2O into peptide domains of PE were calculated as percent of FD PE, observed as the maximum level of PE deuteration (14 h at room temperature, as described in methods). Color coding of deuteration levels from 10 to 90% are indicated in the legend provide with this figure. Mapping of the percent deuteration at the different time points illustrate time-dependent deuteration of PE. Notably, different levels of deuteration were observed for distinct subdomains of PE, illustrated in the PE map. The map shows the primary sequence of recombinant PE with the active enkephalin peptide sequences highlighted in yellow, with the adjacent dibasic residue cleavage sites highlighted in purple.
H-D exchange of intact PE, compared to H-D exchange of peptides derived from PE
For comparison, H-D exchange of intact PE protein was compared with individual peptides derived from PE (by pepsin digestion). The rates of incorporation of deuterium into each peptide fragment present within intact PE, or into individual peptides generated from PE, were plotted (Supporting Information Fig. A). These data indicated differences in relative H-D exchange rates of intact PE compared to its individual peptides, illustrated by mapping such differences throughout PE (Fig. 4). Overall, H-D exchange for intact PE was substantially lower than that for individual peptides derived from PE, illustrating different conformational features of PE compared with peptides of PE. The lower H-D exchange for intact PE suggests that it possesses tertiary conformation that protects numerous subdomains from accessibilities to the aqueous solvent environment.
Figure 4.
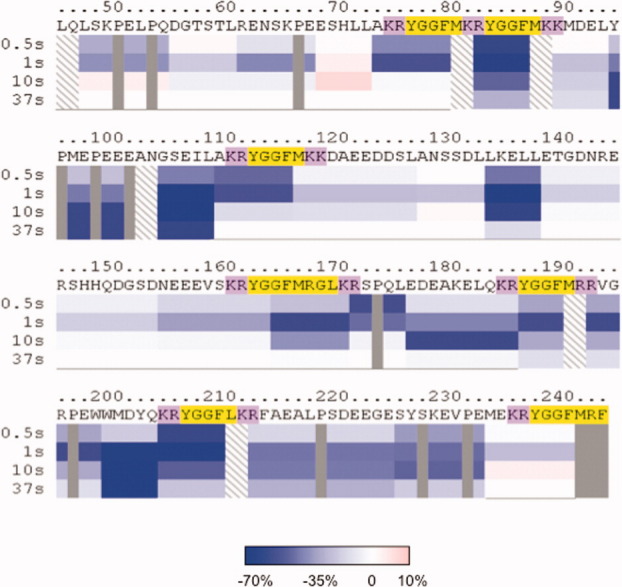
Intact PE displays lower rates of H-D exchange compared to peptides derived from PE. Time-course studies compared the rates of H-D exchange of intact PE compared to individual peptides derived from PE (by pepsin). The percent deuteration of each peptide domain was calculated as the difference between intact PE and peptides derived from PE. The color legend indicates the range of the differences in H-D exchange from −70 to 0% (shades of blue to white) and 0–10% (white to pink). The PE map shows that all the subdomain regions of intact PE have lower H-D exchange rates (shades of blue color) compared to peptides derived from PE.
Differential H-D exchange at proteolytic processing site domains of PE
The multiple protease cleavage sites are essential for processing PE into active enkephalin peptide neurotransmitters and hormones. Therefore, H-D exchange properties of peptide regions spanning the 12 cleavage sites were compared. The relative deuteration of each of the cleavage site “domains” was analyzed for peptide sequences adjacent to the N-terminal and C-terminal sides of the cleavage site that are known to interact with proteases.25, 26 This involved evaluating multiple peptides spanning these cleavage site “domains.”
Differential accessibilities of cleavage site domains of intact PE was observed in time-course studies (0.5 and 1 s incubation in D2O) [Fig. 5(a)] The mid-regions of PE containing cleavage sites #4–7 showed greater deuteration than the adjacent regions of cleavage sites #2–3 and #8–10. However, cleavage sites #1 and #12 at the N- and C-domains of PE demonstrated greater levels of deuteration, similar to the mid-region. Cleavage sites #3, 9, and 10 displayed lower relative deuteration compared with the other protease cleavage sites of PE; the basis for their lower deuteration is not yet known. These H-D exchange data for PE illustrate three main subdomains of PE with differential accessibilities to the aqueous environment corresponding to the mid-region, N- and C-terminal domains, and subdomains between the mid-region and both the N- and C-terminal domains. The cleavage site regions of #4–7 that show high accessibilities (compared with other subdomains of PE) to the aqueous environment in PE represent sites that are readily cleaved by prohormone processing proteases.15, 16, 27
Figure 5.
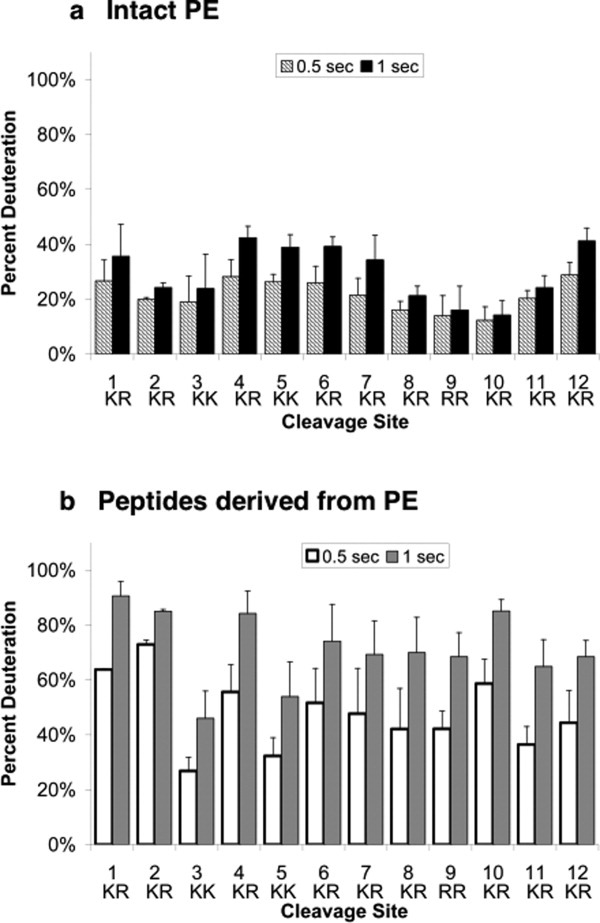
Differential H-D exchange among protease cleavage sites of PE and peptides derived from PE. (a) Intact PE: H-D exchange at cleavage site subdomains. Comparison of relative H-D exchange rates at the 12 dibasic residue cleavage sites of PE are illustrated at 0.5 and 1 s time points for PE incubation in D2O (hatched and black bars, respectively). H-D exchange data was compiled from multiple peptides of PE that span each of the cleavage sites (indicated in figure) and their average level of percent deuteration was calculated (mean ± s.e.m.). (b) Peptides derived from PE: H-D exchange of peptides spanning cleavage site domains. H-D exchange of peptides spanning multiple cleavage sites are illustrated for 0.5 and 1 s exchange time periods (white and gray, respectively). H-D exchange data for peptides spanning each of the cleavage sites (indicated in figure) were compiled and their average level of percent deuteration was calculated (mean ± s.e.m.). The standard deviation of deuterium incorporation measured in replicate determinations was typically less than 5% of the mean, as reported (49, 50).
In contrast to intact PE, peptides derived from PE (by pepsin) showed greater levels of deuteration [Fig. 5(b)]. The majority of individual peptides (derived by pepsin digestion of PE) showed high levels of deuteration at 50–90% of maximum deuteration (at 1 s time point). Peptides at cleavage sites #3 and #5 showed deuteration at about 50% of maximum deuteration. In contrast, intact PE showed lower deuteration throughout the protein at 20–40% of maximum deuteration [1 s time point, Fig. 5(a)]. Direct plots of peptides spanning cleavage sites from DXMS of intact PE, compared with peptides, further illustrate the more limited H-D exchange of peptides of intact PE compared to peptides derived from PE (Supporting Information Fig. B).
Overall, these data illustrate the differential accessibilities of cleavage site domains of PE that undergoes proteolytic processing to generate active enkephalin neurotransmitter peptides.
Discussion
H-D exchange analyses of this study illustrated several unique features of PE protein conformational properties. First, PE undergoes extremely rapid H-D exchange at sub-second and multisecond time points for incubation in heavy water (D2O), which contrasts with many other proteins which display much lower rates of H-D exchange, which include matrilysin, and protein kinase A.17–24 Second, subdomains of the PE prohormone display differences in their relative accessibilities to the aqueous solvent environment, including differences in H-D exchange occurring among the 12 protease cleavage sites of PE. Third, in contrast to intact PE, analyses of individual peptides derived from PE (by pepsin) illustrate their more rapid H-D exchange properties. These data indicate that tertiary conformational properties of PE provide protection for many of its subdomains from accessibilities to the solvent environment, whereas individual peptides (derived from PE) possess full accessibilities to the solvent. In addition, CD (circular dichroism) analyses of PE have demonstrated its secondary structural features composed of α-helix, β-sheet, and β-turns.28 These H-D exchange data indicate that conformational properties of PE participate in its proteolytic processing for production of active enkephalin peptide hormones and neurotransmitters.
It is of interest that the cleavage sites of PE with the greatest accessibilities to the aqueous environment correspond to sites most readily cleaved by processing proteases. Previous studies show initial cleavage of PE at the KR site #7 by cathepsin L,15 a site which shows one of the greatest relative accessibilities to the solvent environment compared with other cleavage sites [Fig. 5(a)]. The prohormone convertases PC1/3 and PC2 show major cleavages of PE at sites #6 and #7, which possess high accessibilities to the aqueous environment16 compared with other cleavage sites. An aspartyl protease involved in prohormone processing readily cleaves site #7 of PE.27 Thus, the DXMS data support the hypothesis that proteolytic processing of PE occurs most readily at cleavage sites with greater accessibilities to the aqueous environment.
It should be noted that in addition to accessibilities of cleavage sites, the amino acid sequences adjacent to the dibasic residue cleavage sites are also important for determining the preferences of proteases for cleavage sites.29–32 Therefore, while the cleavage sites #1, 4, 5, 11, and 12 show similar H-D exchange compared with the most readily cleaved sites #6 and #7, the different amino acid sequences at such sites may participate in the lower extent of their proteolytic processing.15, 16, 27
Differential accessibilities, and thus exchange rates, were observed throughout the PE sequence including cleavage site domains. Interestingly, most segments of intact PE exchanged faster than PE-derived peptides. Areas of more restricted accessibilities were not isolated to cleavage sites. Accessibilities among PE subdomains were varied and specific to particular regions of PE. The rapid H-D exchange periods of sub-second and seconds achieved in this study, compared with longer exchange periods of 100 s in previous studies,28 was key to observing the dynamic nature of PE interactions with its solvent environment. PE represents one among the increasing number of highly dynamic proteins,33 which include protease substrates such as proopiomelanocortin and other similarly dynamic proteins such as α-synuclein.34, 35
The hydrogen-deuterium exchange levels for regions of intact PE and peptides derived from PE were additionally compared with theoretical levels of intrinsic exchange rates for an unstructured polypeptide of the same sequence.36–38 The H-D exchange rates for peptides derived from PE were observed to correlate to the theoretical rates for unstructured peptides of the same sequence. Thus, the more restricted H-D exchange properties of intact PE, occurring rapidly with short exchange periods, indicate its structured features compared with unstructured peptides derived from PE.
These results provide insight into conformational properties of a prohormone, illustrated by H-D exchange investigation of PE in this study. This achievement represents new knowledge of PE and prohormone conformational properties, since no crystal structures of any prohormone has yet been determined. In contrast, crystal structures of the prohormone processing enzyme cathepsin L has been determined,39, 40 and the structural features of the prohormone convertases have been modeled based on furin and kexin crystal structures.41 To gain knowledge of structural features of a prohormone, PE, the DXMS approach of this study illustrates the relative differences in solvent accessibilities of PE subdomains, including its multiple protease cleavage sites.
In conclusion, DXMS studies of PE indicate that (a) PE has protected domains in its tertiary conformation that are relatively protected from the aqueous environment and (b) PE possesses domains with lower solvent accessibility compared to peptides derived from PE. Importantly, the cleavage site domains of PE with the greatest accessibilities to the aqueous environment correspond to sites that are most readily cleaved by prohormone processing proteases.15, 16, 27 It will be important in future studies to gain structural information of PE and prohormones with both DXMS and crystal structure determinations. Such knowledge may promote exploration of small molecules that modulate prohormone interactions with processing proteases as a means for regulating prohormone processing and production of active peptide hormones and neurotransmitters for cell-cell communication in health and disease.
Materials and Methods
Expression of human PE in E. coli and affinity purification
N-His-tagged recombinant human PE was generated by expression in E. coli, and purified by a Ni2+-affinity column as described previously.28 Briefly, PE was expressed using the PE-pET19b vector in the Rosetta 2 (DE3) (EMD, San Diego, CA), BL21 E. coli. After expression, achieved by IPTG induction, PE was purified by Ni2+-affinity column (EMD) equilibrated with 100 mM Na-phosphate, 20 mM Tris, pH 8.0, 6M urea (buffer A), and 5 mM imidazole; PE was eluted with 300 mM imidazole in buffer A and incrementally dialyzed in a buffer representing in vivo reducing conditions in the secretory pathway42 consisting of 100 mM sodium phosphate, 20 mM Tris, pH 7.5, 5 mM glutathione (GSH), and 0.5 mM oxidized glutathione (GSSG; Calbiochem, San Diego, CA). Recombinant PE was dialyzed into a final storage buffer of 50 mM Tris, pH 7.5. Purified recombinant PE was confirmed by SDS-PAGE and immunoblotting with anti-(Met)-enkephalin polyclonal antibody (1:5000, Chemicon, Temecula, CA). Protein concentration was estimated by absorbance measurements at 280 nm using the molar extinction coefficient of 33,640 M−1cm−1 calculated based on the amino acid sequence of recombinant His-tagged PE.43
DXMS studies of intact PE at sub-second and multisecond time points
H-D exchange of intact PE was compared to that of peptides derived from PE obtained by pepsin digestion in time course studies (sub-second and multisecond time points). Each time point used 50 μg of intact PE protein.
H-D exchange of recombinant PE at sub-second and multisecond time points was achieved using the quench-flow apparatus.44 The quench-flow system for these experiments was composed of three Shimadzu LC-10AD HPLC pumps [Fig. 1(b)], with pump A delivering the sample in H2O buffer (8.3 mM Tris, pH 7.2 at 0°C) at 0.1 mL/min and pump B delivering the D2O buffer (8.3 mM Tris in D2O, pH 7.2 at 0°C) at 0.3 mL/min to an Upchurch Scientific M-540 micro static mixing tee (Idex Health & Science, Oak Harbor, WA). Sub-second and multisecond H-D exchange periods were achieved by varying the tubing volume of the delay tube [dashed line in Fig. 1(b)] that delivered the mixture to a second mixing tee where the quenching buffer (1.5M guanidine hydrochloride, 2.4% formic acid (FA), and 24.75% glycerol) was delivered by pump C. H-D exchange experiments were conducted in a water bath of 4.9°C at time points of 0.5, 1, 10, and 37.5 s. After the quenching step, which terminated H-D exchange by decreasing the pH to 2.2–2.5, the sample was collected in a glass microsampling vial (Sun-Sri, Rockwood, TN) and immediately frozen in dry ice. Each time point was collected by manually exchanging the delay tubing [Fig. 1(b)] to the appropriate volume prior to sample collection. The volume within each tube is determined by the inner diameter of the tubing and the length of the tubing. The extent of exposure of the sample to D2O is determined by the volume of that particular tubing because the sample is in continuous flow at 0.4 mL/min prior to being quenched at the second mixing tee.
Each PE sample after H-D exchange was subjected to pepsin digestion and mass spectrometry for analyses of deuterated peptides, conducted as previously described.27, 45, 46 Briefly, the quenched sample at 0°C was passed over a porcine pepsin-immobilized column and the pepsin-generated peptides were separated by a C-18 column (Vydac). The separated peptides were mass analyzed using a Thermo Finnigan LCQ mass spectrometer for identification of pepsin-generated peptide sequences from the resulting MS/MS data sets achieved with use of Sequest (Thermo Scientific, Waltham, MA). Data processing and reduction of H-D exchange experiments used DXMS data-reduction software (Sierra Analytics, Modesto, CA).17, 18, 47
As control, nondeuterated PE was subjected to the same procedure in H2O, allowing analyses of LC-MS/MS spectral data of pepsin-generated fragments of PE. Briefly, the nondeuterated PE samples were processed by addition of 30 μL of 8.3 mM Tris, pH 7.2–10 μL of 50 μg protein or peptides and then the addition of 20 μL of 1.5M guanidine hydrochloride, 2.4% FA, 24.75% glycerol prior to pepsin digestion and LC-MS/MS. In addition, fully deuterated (FD) PE samples had 10 μL of 50 μg protein (or peptides) added to 30 μL of D2O, 0.8% FA and allowed to exchange for 14 h at room temperature prior to pepsin digestion and LC-MS/MS analyses.
Peptides derived from PE by pepsin-digestion and H-D exchange studies
Peptides derived from PE were subjected to H-D exchange for comparison to intact PE. Peptides from PE were generated by initial digestion of PE with pepsin prior to H-D exchange. Pepsin digestion of recombinant PE (2 mg) in 0.5M guanidine hydrochloride, 0.5% FA, 11% glycerol used the pepsin column from H-D exchange experiments described above for intact PE, and as described previously.45, 46 Briefly, the PE protein at 0°C was passed over the porcine pepsin immobilized column using Shimadzu LC-10AD HPLC pumps (Kyoto, Japan) with 0.05% TFA at flow rate of 100 μl/min, and pepsin-generated peptides were directly collected into a test tube on ice. TFA was removed and the peptides were concentrated and isolated using an Amprep™ C-18 mini-column (from Amersham Biosciences, Piscataway, NJ) equilibrated with 2%-acetonitrile in 0.5% FA and elution with 75% acetonitrile, 0.2% FA. The peptides were lyophilized and stored at −70°C prior to use in DXMS experiments. Analyses of peptides derived from 50 μg PE was used for each H-D exchange experiment, conducted as described above for intact PE. Lyophilized peptides were reconstituted in 8.3 mM Tris, pH 7.2 prior to H-D exchange and LC-MS/MS.
Analyses of H-D exchange rates
The exchange process in a protein/peptide of x number of amides can be viewed as x number of independent chemical reactions that each obeys first order reaction kinetics.36–38, 48 If amide hydrogen i exchanges at the rate of kex,i, the amount of deuterium of this position at time point t could be computed as Di,t = 1 − e−kex,it. kex,i, is a function of pH, temperature, protein sequence, and protein conformation. The exchange rate of each peptide or segments of protein is a summation of the H-D exchange for each amide hydrogen, with the exceptions of the N-terminal amino group of the peptide and the amide hydrogen of the second amino acid, which exchange too rapidly to retain deuterons during experiments, and the amino acid proline, which does not contain an amide proton. Therefore, for a fragment f starting from residue m to n, the amount of deuterium incorporated is 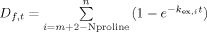 .
.
A double exponential equation, D = Amax − A1e−k1t − A2e−k2t, 41 was used to fit the sub-second H-D exchange data and calculate the H-D exchange rates using MatLab (MathWorks, Natick, MA) and Mathematica 6.0 (Wolfram Research, Champaign, IL). The Amax term was used as a constraint of A1 and A2 such that the sum is less than or equal to the maximum hydrogen-deuterium exchangeable sites (A1 + A2 = Amax). The resulting parameters were then used in the comparison of the exchange rates between intact PE and peptides derived from PE (Fig. 4). In generating the ribbon maps of Figures 3 and 4, the relative deuteration levels of the peptides were sublocalized using overlapping peptides.45 First, fragments (fi) were delineated within each peptide based on the overlapping regions with another peptide or set of peptides. To prevent over interpretation of minimally overlapping regions, a minimum overlap of five residues was established. Second, we defined a variable si,t to represent the mass shift of fragment fi at one exchange time point t. And thus, a set of linear equations was established such that the sum of si,t in a specific peptide was equal to the total number of experimentally observed deuterium for that particular peptide. A best fit solution was obtained by linear least square fitting, with the following two constraints: (1) si,t could not be <0 and (2) for a given fragment fi, mass shift at longer on-exchange time points could not be smaller than mass shift at the shorter on-exchange time points. These fitted values were then plotted as shown in Figure 5.
Glossary
Abbreviations
- AcN
acetonitrile
- DXMS
hydrogen-deuterium exchange mass spectrometry
- FA
formic acid
- H-D exchange
hydrogen-deuterium exchange
- HPLC
high pressure liquid chromatography
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- PE
proenkephalin
- TFA
trifluoroacetic acid
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Docherty K, Steiner DF. Post-translational proteolysis in polypeptide hormone biosynthesis. Annu Rev Physiol. 1982;44:625–638. doi: 10.1146/annurev.ph.44.030182.003205. [DOI] [PubMed] [Google Scholar]
- 3.Holden JE, Jeong Y, Forrest JM. The endogenous opioid system and clinical pain management. AACN Clin Issues. 2005;16:291–301. doi: 10.1097/00044067-200507000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Kieffer BL, Gavériaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 5.Terenius L. From opiate pharmacology to opioid peptide physiology. Ups J Med Sci. 2000;105:1–15. doi: 10.1517/03009734000000043. [DOI] [PubMed] [Google Scholar]
- 6.Meilandt WJ, Yu GQ, Chin J, Roberson ED, Palop JJ, Wu T, Scearce-Levie K, Mucke L. Enkephalin elevations contribute to neuronal and behavioral impairments in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2008;28:5007–5017. doi: 10.1523/JNEUROSCI.0590-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamei J, Miyata S, Takahashi M, Saitoh A. Involvement of delta1-opioid receptors in the spatial learning impairment in streptozotocin-induced diabetic mice. Nihon Shinkei Seishin Yakurigaku Zasshi. 2005;25:221–225. [PubMed] [Google Scholar]
- 8.Nitsche JF, Schuller AG, King MA, Zengh M, Pasternak GW, Pintar JE. Genetic dissociation of opiate tolerance and physical dependence in delta-opioid receptor-1 and preproenkephalin knock-out mice. J Neurosci. 2002;22:10906–10913. doi: 10.1523/JNEUROSCI.22-24-10906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shippenberg TS, LeFevour A, Chefer VI. Targeting endogenous mu- and delta-opioid receptor systems for the treatment of drug addiction. CNS Neurol Disord Drug Targets. 2008;7:442–453. doi: 10.2174/187152708786927813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berrendero F, Robledo P, Trigo JM, Martín-García E, Maldonado R. Neurobiological mechanisms involved in nicotine dependence and reward: participation of the endogenous opioid system. Neurosci Biobehav Rev. 2010;35:220–231. doi: 10.1016/j.neubiorev.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci USA. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang SR, Garza C, Mosier C, Toneff T, Wunderlich E, Goldsmith P, Hook V. Cathepsin L expression is directed to secretory vesicles for enkephalin neuropeptide biosynthesis and secretion. J Biol Chem. 2007;282:9556–9563. doi: 10.1074/jbc.M605510200. [DOI] [PubMed] [Google Scholar]
- 13.Seidah NG, Prat A. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 2002;38:79–94. doi: 10.1042/bse0380079. 2002. [DOI] [PubMed] [Google Scholar]
- 14.Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 15.Schiller MR, Mende-Mueller L, Moran K, Meng M, Miller KW, Hook VYH. “Prohormone thiol protease” (PTP) processing of recombinant proenkephalin. Biochemistry. 1995;34:7988–7995. doi: 10.1021/bi00025a004. [DOI] [PubMed] [Google Scholar]
- 16.Breslin MR, Lindberg I, Benjannet S, Mathis JP, Lazure C, Seidah NG. Differential processing of proenkephalin by prohormone converteases 1(3) and 2 and furin. J Biol Chem. 1993;268:27084–27093. [PubMed] [Google Scholar]
- 17.Englander JJ, Del Mar, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods VL. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL. Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia RA, Pantazatos DP, Gessner CR, Go KV, Woods VL, Villarreal FJ. Molecular interactions between matrilysin and the matrix metalloproteinase inhibitor doxycycline investigated by deuterium exchange mass spectrometry. Mol Pharmacol. 2005;67:1128–1136. doi: 10.1124/mol.104.006346. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Tsalkova T, White MA, Mei FC, Liu T, Wang D, Woods VL, Jr, Cheng X. Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS) J Biol Chem. 2011;286:17889–17897. doi: 10.1074/jbc.M111.224535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamuro Y, Burns LL, Canaves JM, Hoffman RC, Taylor SS, Woods VL. Domain organization of D-AKAP2 revealed by enhanced deuterium exchange-mass spectrometry (DXMS) J Mol Biol. 2002;321:703–714. doi: 10.1016/s0022-2836(02)00419-9. [DOI] [PubMed] [Google Scholar]
- 22.Hamuro Y, Wong L, Shaffer J, Kim JS, Stranz DD, Jennings PA, Woods VL, Adams JA. Phosphorylation driven motions in the COOH-terminal Src kinase, Csk, revealed through enhanced hydrogen -deuterium exchange and mass spectrometry (DXMS) J Mol Biol. 2002;323:871–881. doi: 10.1016/s0022-2836(02)01003-3. [DOI] [PubMed] [Google Scholar]
- 23.Hamuro Y, Anand GS, Kim JS, Juliano C, Stranz DD, Taylor SS, Woods VL. Mapping intersubunit interactions of the regulatory subunit (RI alpha) in the type I holoenzyme of protein kinase A by amide hydrogen/deuterium exchange mass spectrometry (DXMS) J Mol Biol. 2004;340:1185–1196. doi: 10.1016/j.jmb.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 24.Spraggon G, Pantazatos D, Klock HE, Wilson IA, Woods VL, Lesley SA. On the use of DXMS to produce more crystallizable proteins: structures of the T. maritima proteins TM0160 and TM 1171. Protein Sci. 2004;13:3187–3199. doi: 10.1110/ps.04939904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 26.Schechter I, Berger A. On the active site of proteases III. Mapping the active site of papain; Specific peptide inhibitors of papain. Biochem Biophys Res Commun. 1968;32:898–902. doi: 10.1016/0006-291x(68)90326-4. [DOI] [PubMed] [Google Scholar]
- 27.Azaryan AV, Schiller M, Mende-Mueller L, Hook VYH. Characteristics of the chromaffin granule aspartic proteinase involved in proenkephalin processing. J Neurochem. 1995;65:1771–1779. doi: 10.1046/j.1471-4159.1995.65041771.x. [DOI] [PubMed] [Google Scholar]
- 28.Lu WD, Asmus K, Hwang SR, Li S, Woods VL, Hook V. Differential accessibilities of dibasic prohormone processing sites of proenkephalin to the aqueous environment revealed by H-D exchange mass spectrometry. Biochemistry. 2009;48:1604–1612. doi: 10.1021/bi801888j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schechter I. Mapping of the active site of proteases in the 1960s and rational design of inhibitors/drugs in the 1990s. Curr Protein Pept Sci. 2005;6:501–512. doi: 10.2174/138920305774933286. [DOI] [PubMed] [Google Scholar]
- 30.Schilling S, Kohlmann S, Bäuscher C, Sedlmeier R, Koch B, Eichentopf R, Becker A, Cynis H, Hoffmann T, Berg S, Freyse EJ, von H, rsten S, Rossner S, Graubner S, Demuth HU. Glutaminyl cyclase knock-out mice exhibit slight hypothyroidism but no hypogonadism: implications for enzyme function and drug development. J Biol Chem. 2011;286:14199–14208. doi: 10.1074/jbc.M111.229385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su YC, Miller TN, Navaneetham D, Schoonmaker RT, Sinha D, Walsh PN. The role of factor XIa (FXIa) catalytic domain exosite residues in substrate catalysis and inhibition by the Kunitz protease inhibitor domain of protease nexin 2. J Biol Chem. 2011;286:31904–31914. doi: 10.1074/jbc.M111.257527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosseini M, Jiang L, Sørensen HP, Jensen JK, Christensen A, Fogh S, Yuan C, Andersen LM, Huang M, Andreasen PA, Jensen KJ. Elucidation of the contribution of active site and exosite interactions to affinity and specificity of peptidylic serine protease inhibitors using non-natural arginine analogs. Mol Pharmacol. 2011;80:585–597. doi: 10.1124/mol.111.072280. [DOI] [PubMed] [Google Scholar]
- 33.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 35.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL. Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 37.Bai YW, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen-exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujishima A, Imai Y, Nomura T, Fujisawa Y, Yamamoto Y, Sugawara T. The crystal structure of human cathepsin L complexed with E-64. FEBS Lett. 1997;407:47–50. doi: 10.1016/s0014-5793(97)00216-0. [DOI] [PubMed] [Google Scholar]
- 40.Coulombe R, Grochulski P, Sivaraman J, Ménard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–503. [PMC free article] [PubMed] [Google Scholar]
- 41.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J Mol Biol. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 42.Yasothornsrikul S, Aaron W, Toneff T, Hook VY. Evidence for the proenkephalin processing enzyme prohormone thiol protease (PTP) as a multicatalytic cysteine protease complex: activation by glutathione localized to secretory vesicles. Biochemistry. 1999;38:7421–7430. doi: 10.1021/bi990239w. [DOI] [PubMed] [Google Scholar]
- 43.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 44.Rist W, Rodriguez F, Jorgensen TJ, Mayer MP. Analysis of subsecond protein dynamics by amide hydrogen exchange and mass spectrometry using a quenched-flow setup. Protein Sci. 2005;14:626–632. doi: 10.1110/ps.041098305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL. Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14:2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derunes C, Burgess R, Iraheta E, Kellerer R, Becherer K, Gessner CR, Li S, Hewitt K, Vuori K, Pasquale EB, Woods VL, Ely KR. Molecular determinants for interaction of SHEP1 with Cas localize to a highly solvent-protected region in the complex. FEBS Lett. 2006;580:175–178. doi: 10.1016/j.febslet.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 47.Pantazato D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL. Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci USA. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 49.Mendillo ML, Putnam CD, Mo AO, Jamison JW, Li S, Woods VL, Jr, Kolodner RD. Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. J Biol Chem. 2010;285:13170–13182. doi: 10.1074/jbc.M110.108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu S, Kim Y, Li S, Durrant ES, Pace RM, Soods VL, Gentry MS. Structural insights into glucan phosphatase dynamics using amide hydrogen-deuterium exchange mass spectrometry. Biochemistry. 2009;48:9891–9902. doi: 10.1021/bi9008853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.