Figure 1. A diagram showing miRNAs and their targets in cardiac hypertrophy.
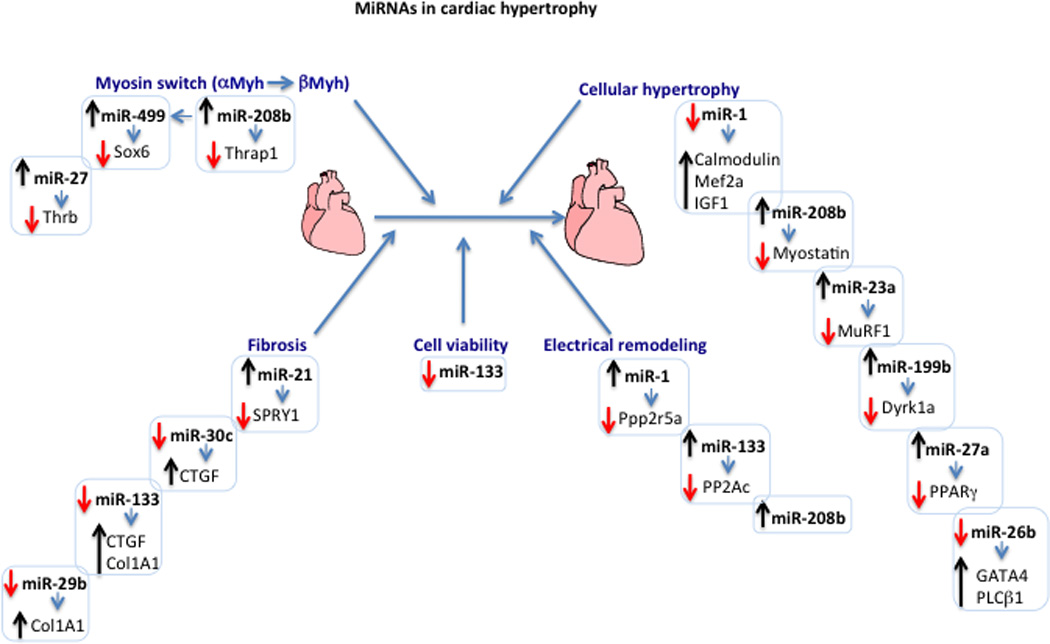
The diagram displays the different miRNAs and their targets that are involved in gene switching, cellular hypertrophy, fibrosis, and electrical remodeling during cardiac hypertrophy. Upregulation or downregulation of a specific miRNA is represented by an upward (black) or a downward (red) arrow, respectively. The change in the expression levels of target genes inversely correlates with that of the targeting miRNA and is similarly represented by an up or down arrow. All listed targets have been validated. The listed targets include: thyroid hormone receptor-associated protein 1 (Thrap1), SRY-box containing gene 6 (Sox6), thyroid hormone receptor beta (Thrb), calmodulin, myocyte enhancer factor 2A (Mef2a), insulin-like growth factor 1 (IGF1), myostatin, muscle-specific RING finger protein 1 (MuRF1), dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (Dyrk1a), sprouty 2/4 (SPRY2/4), connective tissue growth factor (CTGF), collagen IA1 (Col1A1), GATA binding protein 4 (GATA4), phospholipase C beta 1 (PLCβ1), protein phosphatase 2, regulatory subunit B (B56) alpha isoform (Ppp2r5a), protein phosphatase 2 catalytic subunit (PP2Ac) and, the indirect targets alpha and beta myosin heavy chain (αand βMyh),
