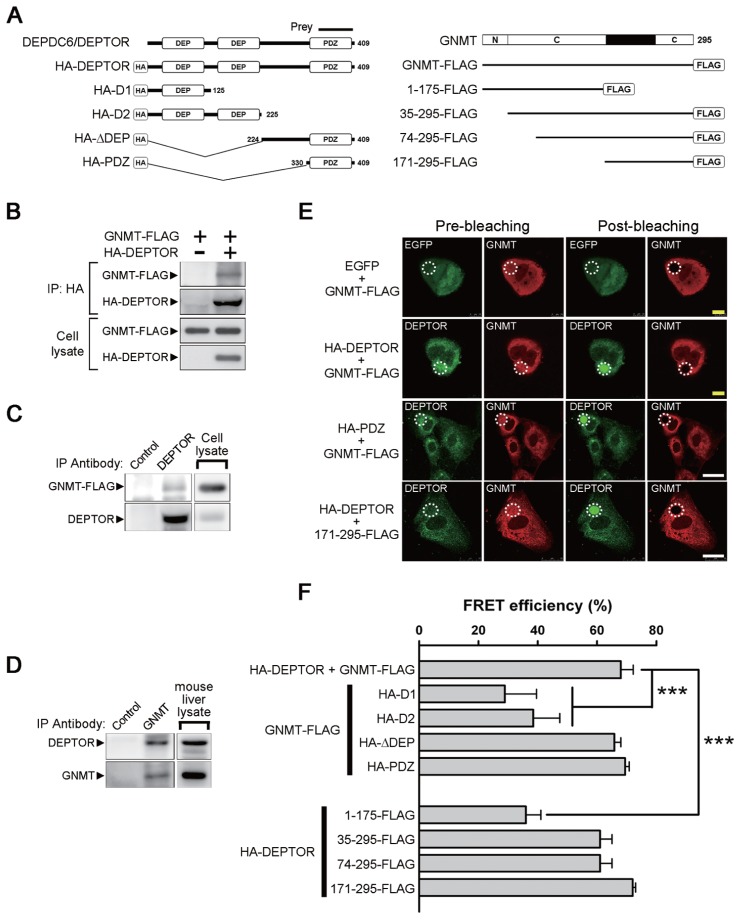
Figure 1.
Identification of DEPDC6/DEPTOR as a GNMT binding protein and mapping of the interactive domains. (A) Different constructs of DEPDC6/DEPTOR and GNMT used for domain mapping. The protein fragment expressed by the positive prey clone obtained from a yeast two-hybrid screening was indicated. N, amino-terminal region of GNMT; C, catalytic domains located in amino acids 37–175 and 243–295 of GNMT. (B) FLAG-tagged GNMT coimmunoprecipitated with HA-tagged DEPTOR. HEK293T cells were transfected with the indicated plasmids and harvested for immunoprecipitation analysis. (C) Interaction of endogenous DEPTOR with FLAG-tagged GNMT. FLAG-tagged GNMTs were expressed in HuH-7 cells, and cell lysates were used for immunoprecipitation analysis. (D) Interaction of endogenous DEPTOR with endogenous GNMT. Mouse liver lysates were used for immunoprecipitation analysis. Each experiment was repeated at least three times. (E) FRET-AB assay was used to assess the interaction between GNMT and DEPTOR and to map their interactive domains. Photobleaching of the GNMT rhodamine label (white dotted circle) resulted in an increase of DEPTOR fluorescent signal within the photobleached area, thus demonstrating the FRET effects (yellow bar = 10 μm; white bar = 25 μm). Positive results were also observed in HuH-7 cells coexpressed in the PDZ domain of DEPTOR and GNMT-FLAG as well as in cells coexpressed in the HA-DEPTOR and C-terminal region (amino acids 171–295) of GNMT. (F) Quantitative results of FRET efficiency in different sets of FRET-AB assays. HuH-7 cells were transfected with the indicated plasmids and were subjected to FRET-AB assay. Data are presented as means ± SD of 5–10 random fields from two independent experiments. Some image data are shown in Supplementary Figure 2. ***P < 0.001. IP, immunoprecipitation.