Abstract
The silver carp (Hypophthalmichthys molitrix) is among the most intensively pond-cultured fish species and is used in the wild to counteract water bloom in China. However, little genomic information is available for this species, especially regarding its ability to grow rapidly in water, even water contaminated with high concentrations of poisonous microcystin. In this study, we performed de novo transcriptome assembly and analysis of the 17.10 million short-read sequences produced by the Illumina paired-end sequencing technology. Using an improved multiple k-mer contig assembly method coupled with further scaffolding, 85 759 sequences were obtained. There were 23 044 sequences annotated with 3423 gene ontology terms for 104 196 term occurrences and the three corresponding organizing principles. A total of 38 200 assembled sequences were involved in 218 predicted Kyoto Encyclopedia of Genes and Genomes metabolic pathways. We also recovered 41 of 44 genes involved in the biosynthesis of glutathione. Of these, five genes were identified as experienced positive selection between silver carp and zebrafish, as determined by the likelihood ratio test. This report is the first annotated review of the silver carp transcriptome. These data will be of interest to researchers investigating the evolution and biological processes of the silver carp. This work also provides an archive for future studies of recent speciation and evolution of Cyprinidae fishes and can be used in comparative studies of other fishes.
Keywords: silver carp, transcriptome, paired-end, RNA sequencing
1. Introduction
The transcriptome is made up of the subset of genes active in a selected tissue and species. Understanding the dynamics of the transcriptome is essential for interpreting phenotypic variation caused by combinations of genotypic and environmental factors.1 Massively parallel sequencing of RNA2 (RNA-Seq) has offered the opportunity to characterize the transcriptome with unprecedented sensitivity and depth. It has already revolutionized the way we study the transcriptome. The latest paired-end sequencing of RNA-Seq techniques have further improved the efficiency of DNA sequencing and expanded short read lengths, permitting a deeper understanding of the transcriptome.3 RNA-Seq is independent of prior knowledge and does not require design work, thus reducing the required staff, cost and time and providing the unprecedented opportunity to conduct low-cost transcriptome studies at lower cost for non-model organisms. The RNA-Seq technology has been applied to many model organisms4–12 for the discovery of splice variants, RNA editing sites and new microRNAs, but fewer studies were conducted in non-model fish organisms.13,14
The Actinopterygii, in terms of numbers, are the dominant class of vertebrate, comprising nearly 96% of the 26 000 species of fish.15 However, the genomic information of this group is very rare: only six genomes16–19 and several transcriptomes20,21 are available. This has hindered research into these valuable species. Cyprinidae is the largest family of freshwater fish in the Actinopterygii.15 The endemic clade of East Asian Cyprinidae displays a tremendous diversity of phenotypic and ecological traits in this area. This clade is an ideal model system for the study of rapid radiations and evolutionary adaptation over short periods of time.22 Silver carp (Hypophthalmichthys molitrix) of the family Cyprinidae are among the most intensively pond-cultured species in China. As one of famous four major Chinese carps, breeding production reached 3 million tons in China in 2009.23 Aside from their great importance to the fishery economy, silver carp have been found useful in counteracting cyanobacteria blooms in China.24,25 Silver carp are also a good model for the study of speciation because of its split with bighead carp (Hypophthalmichthys nobilis), which occurred only ∼3 Mya.26 However, lack of genomic resources like genome sequence, transcriptome sequences and molecular markers has made the study of silver carp breeding, the mechanism of its ability to counteract water bloom and evolutionary analysis a difficult task.
When no genome sequence is available, transcriptome sequencing is an effective way to obtain large numbers of molecular makers and identify transcripts involved in specific biological processes. In this study, we present the first silver carp transcriptome using massively parallel mRNA sequencing. We perform Illumina sequencing of the heart, liver, brain, spleen and kidney tissues to characterize the H. molitrix transcriptome. A database (Silver Carp Base) is under construction and we expect that it will provide the first picture of the transcriptome of this species. The database will be updated in the future if additional data become available.
2. Materials and methods
2.1. Ethics statement
All experimental protocols were approved by the ethics committee of Institute of HydroBiology, Chinese Academy of Sciences.
2.2. Organ collection and RNA isolation
A wild silver carp was collected from the middle reach of the Yangtze River. To obtain the whole transcriptome, RNA from five organs (heart, liver, brain, spleen and kidneys) was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After the quality examination by the way of electrophoresis and a BioPhotometer plus 6132 (Eppendorf, Germany), RNAs from different organs were mixed together at equivalent concentrations. Total RNA extraction was in accordance with the manufacturer's protocol and it was treated with RNase-free DNase I (New England Biolabs) for 30 min at 37°C to remove the residual DNA.
2.3. cDNA library preparation and sequencing
Beads with oligo(dT) were used to purify poly(A) mRNA from total RNA. Then, the mRNA was fragmented using a RNA fragmentation kit (Ambion). First-strand cDNA was synthesized using random hexamer-primer and reverse transcriptase (Invitrogen), and second-strand cDNA was synthesized next. Then the paired-end cDNA library was prepared in accordance with Illumina's protocols with an insert size of 200 bp and sequenced for 75 bp. The Illumina GA processing pipeline v0.2.2.6 was used to analyze the image and for base calling.
2.4. De novo assembly of silver carp transcriptome
As no optimal k-mer length is appropriate for all de novo transcriptome assemblies, the multiple k-mer method was used to obtain longer silver carp mRNA sequences, which are very useful in subsequent analysis steps. Our method is based on the modified ‘additive Multi-k’ method described by Yann Surget-Groba27 After removing reads with the sequencing adapter and reads of low quality, paired-end reads were subjected to de novo assembly using ABySS28 with k-mer lengths of 58, 54, 52, 50, 48, 46, 44, 42, 40, 38 and 34. The unused reads at higher k-mer lengths were not discarded before running the assembly for a lower k-mer length. The output data set of each k-mer length was subjected to SSPACE29 for scaffolding, respectively. When pooling all the results together, some contigs and scaffolds appeared in two or more assemblies, causing redundancy. These were removed using CD-HIT-EST.30 The longest possible contigs and scaffolds were retained. At last, the STM+ method27 was used to perform translation mapping scaffolding with the Danio rerio proteome31 serving as a reference.
2.5. Sequence annotation
The assembled sequences were blasted against the NCBI Nr (non-redundant) protein database and Swiss-prot database using BLASTX32 and an E-value of 1e−5. To shorten the search time, searches were limited to the first 10 significant hits for each query. Gene names were assigned to each sequence according to its best BLAST hit (highest score).
The Blast2GO suit33 was used for functional annotation of assembled sequences applying the function for the mapping of gene ontology (GO) terms to sequences with BLAST hits obtained from hits with E-value < 1e−5, annotation cut-off > 55 and a GO weight > 5 were used for annotation. Assembled sequences were thus assigned to primary and sub-GO functional categories.
2.6. Simple sequence repeat markers discovery
A microsatellite program (MISA)34 (http://pgrc.ipkgatersleben.de/misa/) was used to identify and localize microsatellite motifs. We searched for all types of simple sequence repeats (SSRs) from mononucleotide to hexanucleotides using the following parameters: at least 10 repeats for mono-, 6 repeats for di- and 5 repeats for tri-, tetra-, penta- and hexanucleotide for simple repeats. Both perfect (i.e. SSRs contain a single repeat motif like such as ‘ATC’) and compound (i.e. composed of two or more motifs separated by <100 bp) SSRs were identified.
2.7. Positive selection
All the 44 sequences involved in the biosynthesis of glutathione (GSH) were downloaded from Kyoto Encyclopedia of Genes and Genomes (KEGG) and used as a query to blast against the 85 759 sequences assembled. Only the reciprocal BLAST best-hit result sequence was kept to form the sequence pair with its corresponding query. To determine whether the sequence pair underwent positive selection, a likelihood ratio test (LRT) was performed for the Nsite 7 and Nsite 8 of codeml in PAML was used.35
3. Results
3.1. De novo assembly with multiple k-mer lengths and sequence validation
We prepared the mixed cDNAs from the heart, liver, brain, spleen and kidneys of silver carp at equivalent concentration. One lane of Illumina Genome Analyzer was performed and ∼17.10 million 75 bp paired-end reads were obtained. After cleaning the low-quality reads, we used a modified version of a previous published procedure27 (see Materials and methods) to assemble the reads for non-redundant consensus. The bioinformatics workflow is depicted in the flowchart shown in Supplementary Fig. 1S. Short read data have been deposited in NCBI's Short Read Archive at http://www.ncbi.nlm.nih.gov/sra under the accession SRP008133.
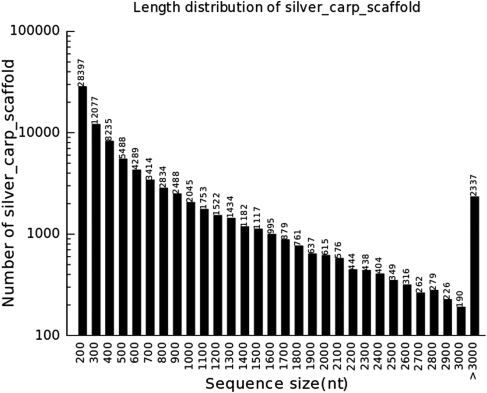
To assemble the paired-end reads into contigs, we used ABySS28 with different k-mer lengths (Table 1). Although paired-end information has been used in ABySS, a great improvement was found after scaffolding the contigs with SSPACE29 (Table 2). After pooling all the scaffolds obtained from multiple k-mers together, 3 930 925 sequences were collected. Using CD-HIT-EST,30 scaffolds were assembled into clusters that were analyzed for consensus. Finally, 85 796 sequences ranging from 200 to 13 880 bp were collected. The length distribution of all the sequences is shown in Fig. 1.
Table 1.
Summary statistics of the assemblies used to assess the performances of the Mulit-K de novo assembly method
| Method | k-mer | Contig > 100 | N50 | Max length | Total length (Mb) | Average contig size |
|---|---|---|---|---|---|---|
| single K | 58 | 3328 | 65 | 3324 | 2.076 | 87 |
| 54 | 22 397 | 159 | 5597 | 8.197 | 127 | |
| 52 | 37 207 | 233 | 6087 | 12.602 | 140 | |
| 50 | 51 041 | 239 | 8297 | 18.059 | 142 | |
| 48 | 61 097 | 241 | 8045 | 23.245 | 144 | |
| 46 | 69 717 | 242 | 8358 | 28.235 | 145 | |
| 44 | 77 038 | 239 | 11 062 | 33.133 | 142 | |
| 42 | 82 806 | 236 | 10 004 | 37.633 | 139 | |
| 40 | 87 673 | 233 | 7322 | 41.928 | 135 | |
| 38 | 91 694 | 228 | 10 092 | 45.964 | 130 | |
| 34 | 97 153 | 220 | 13 873 | 53.206 | 119 | |
| multi K | 118 764 | 257 | 13 880 | 58.075 | 159 |
These statistics correspond to the set of contig > 100 bp. k-mer, required length of identical overlap match between two reads by ABySS; N50, contig length–weighted median; max length, length of the longest contig; (Total length) summed length of all contig > 100 bp.
Table 2.
Summary statistics of the scaffolds produced by SSPACE
| k-mer | scaffold > 100 | N50 | Max length | Total length (Mb) | Average scaffold size |
|---|---|---|---|---|---|
| 58 | 2805 | 65 | 4835 | 2.074 | 92 |
| 54 | 19 184 | 241 | 7069 | 8.165 | 135 |
| 52 | 31 314 | 279 | 11 041 | 12.561 | 151 |
| 50 | 41 815 | 301 | 11 669 | 18.033 | 155 |
| 48 | 49 814 | 325 | 12 403 | 23.239 | 159 |
| 46 | 57 241 | 332 | 10 964 | 28.259 | 160 |
| 44 | 63 799 | 324 | 11 062 | 33.196 | 156 |
| 42 | 69 856 | 314 | 10 950 | 37.804 | 152 |
| 40 | 74 827 | 302 | 12 140 | 42.046 | 146 |
| 38 | 79 408 | 291 | 11 339 | 46.097 | 139 |
| 34 | 87 408 | 270 | 13 880 | 53.831 | 127 |
These statistics correspond to the set of scaffold > 100 bp. k-mer, required length of identical overlap match between two reads by ABySS; N50, scaffold length–weighted median; max length, length of the longest scaffold; total length, summed length of all scaffold > 100 bp.
Figure 1.
Length distributions of scaffolds assembled by a multiple k-mer method.
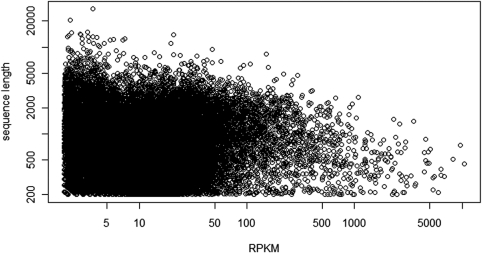
To determine the expression level of the transcripts, we mapped the raw reads to the assembled sequences with SOAP36 and the RPKM value (Reads Per Kilobase of exon model per Million mapped reads) of all the transcripts are shown in Supplementary Table S1. Figure 2 depicts the relationship of RPKM versus the transcript size. Transcript length increased with coverage depth and reached an asymptote approximately at an average coverage of ∼50.
Figure 2.
The relationship of RPKM versus the transcript size. RPKM, Reads Per Kilobase of exon model per Million mapped reads.
Until now, no general criteria have been proposed as standards for evaluation of the quality of transcriptome assembly. We used three substantial factors to assess how well the assembled sequences represent the actual transcriptome population: (i) gene coverage, (ii) transcript sequence quality and (iii) completeness.
The transcriptome gene coverage was judged by comparison with the sequence information available for silver carp. All 13 mitochondrial protein-coding genes and 203 of 217 proteins in the NCBI database were present in our assembled scaffolds. We compared our assembled scaffolds with the zebrafish transcriptome (ENSEMBL Zv61) and found that 40 509 of 41 759 (85.9%) zebrafish transcripts have matches in assembled scaffolds. At the same time, 19 893 reciprocal best-hit blast matches with the zebrafish transcriptome were identified using E-value 1e−5.
Transcriptome quality was assessed by comparing the mitochondrial protein-coding genes found in assembled sequences to mitochondrion sequence in GenBank (NC_010156). A total of 10 185 nucleotide identities were observed out of 10 522 (96.8%) total nucleotide length of contig to coding mitochondrial sequences BLAST matches, suggesting very good transcriptome sequence quality. The observed 3.2% sequence difference might be due to the high intraspecific genetic variability.
Finally, in terms of sequence completeness, the relative number of full-length sequences in the 19 893 reciprocal best-hit blast matches to zebrafish transcriptome was estimated. A sequence was considered full length if it contained the complete 5′- and 3′-UTR of the mRNA. In this study, we used a less stringent but broadly adopted definition, considering a sequence to be full length if it comprised at least the complete coding sequence (CDS).21 We mapped the 19 893 sequences to their corresponding CDS in the zebrafish transcript, and if the CDS was fully covered by assembled sequence, we thought the sequence as full-length sequence. Under the criteria given above, 1937 sequences (9.7%) were validated as full length. One thousand, six hundred and thirty-five sequences (8.2%) covered more than 95% of the zebrafish CDS and 2394 sequences (12.0%) covered more than 75% of the zebrafish CDS. For a pseudo-stop codon usually appears on a chimeric or truncated transcript, we translated the nucleotide sequences to protein sequences to verify the completeness of the transcripts. One thousand, three hundred and seventy-seven sequences were validated as full length and 2109 sequences covered more than 95% of the zebrafish CDS.
In addition to the computing methods given above, Reverse transcription polymerase chain reaction (RT-PCR) assay was used to validate the quality of the assembled transcriptome. Primers for 22 transcripts with different expression levels (RPKM ranged from 336 to 10 507) were designed and all the cDNAs were amplified. Out of the 22 pairs of primers, 10 pairs were silver-specific transcripts which did not have an NCBI Nr BLAST hit (see Sequence annotation). The primer information and RT–PCR results are shown in Supplementary Table S2 and Fig. S2.
3.2. Sequence annotation
Several complementary methods were used to annotate the assembled sequences. First, the assembled sequences were searched against the Nr protein databases using BLASTX with an E-value of 1e−5. Of the 85 796 assembled sequences, 54 198 (63.2%) had significant matches (Supplementary Table S3). Most of the sequences with top-hit blast result from zebrafish (44 999 sequences; 83.0%). In addition, 18 536 (34.2%) sequences matched predicted proteins, 81 (0.1%) with unknown proteins.
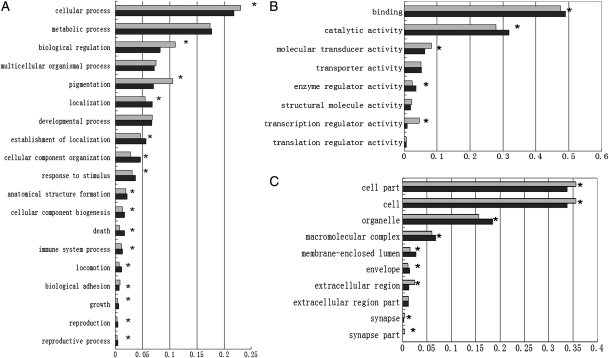
Second, silver carp sequences that had matches in Nr databases were given GO annotations with the Uniprot database. Of these, 23 044 were assigned to one or more 3423 GO terms, for a total of 104 196 term occurrences. As many as 17 451 sequences were found to be involved in biological process and could be divided into cellular process (13 382 sequences with percentage of 76.7%), metabolic process (10 846; 62.2%), biological regulation (5032; 28.9%), multicellular organismal process (4386; 25.1%), pigmentation (4296; 24.6%), localization (4162; 23.8%), developmental process (4088; 23.4%), establishment of localization (3448; 19.8%), cellular component organization (2793; 16.0%) and response to stimulus (2275; 13.0%). Other type of functions occurred at <10% each (Fig. 3).
Figure 3.
Functional classification of silver carp transcriptome and comparison with zebrafish transcriptome. (A) GO: biological process. (B) GO: molecular function. (C) GO: cellular component. In some cases, one transcript may have multiple functions. Grey, silver carp; black, zebrafish.
GO analysis have also shown that 15 799 sequences were associated with a cellular component, including cell (15 114; 95.6%), cell part (15 114; 95.6%), organelle (8265; 52.3%), organelle part (3624; 22.9%) and macromolecular complex (3042; 19.2%). Moreover, 17 837 sequences showed potential molecular function, such as binding (13 027; 73%), catalytic activity (8473; 47.5%), molecular transducer activity (1648; 9.2%) and transporter activity (1406; 7.8%). The detailed information about the functional classification is shown in Supplementary Table S3.
Representation of GO categories in the silver carp transcriptome set was found to be similar to that of the zebrafish GO database, but there were a few differences in each of the three main GO categories (Fig. 3). After correcting for multiple tests, we found that 30 of 37 comparisons were significantly over or underrepresented in comparison to the zebrafish records. For example, among the biological processes, pigmentation (GO: 0043473) was underrepresented in the silver carp transcriptome, while localization (GO: 0051179) and response to stimulus (GO: 0050896) were overrepresented.
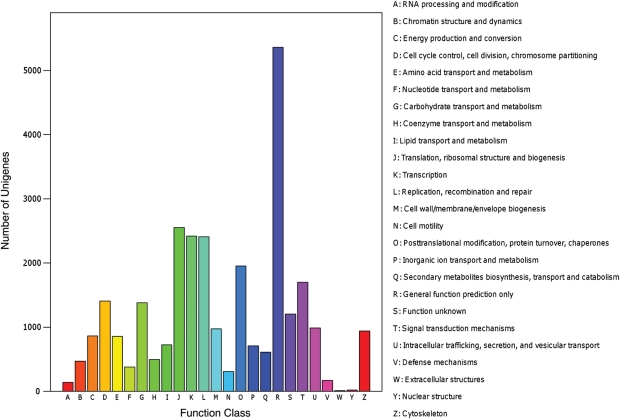
Meanwhile, annotation of the 85 759 sequences using Clusters of Orthologous Groups of protein (COG) databases yielded good results for 14 840 putative proteins. The COG-annotated putative proteins ranged functionally into at least 25 molecular families, including biochemical metabolism, signal transduction, cellular structure and immune defense, in accordance with the categories observed in GO annotation (Fig. 4).
Figure 4.
COG annotations of putative proteins. All putative proteins were aligned to COG database and can be classified functionally into at least 25 molecular families.
3.3. Metabolic pathways by KEGG analysis
A total of 38 200 assembled sequences were found to be involved in 218 predicted KEGG metabolic pathways. The number of sequences ranged from 3 to 4510 (Supplementary Table S4). The top 20 pathways with the greatest number of sequences are shown in Table 3, and the greatest number of transcripts was found in the metabolic pathways. The top 10 metabolic pathways were: purine metabolism (789), pyrimidine metabolism (473), oxidative phosphorylation (436), inositol phosphate metabolism (435), glycerophospholipid metabolism (371), riboflavin metabolism (347), glycolysis/gluconeogenesis (341), lysine degradation (337), pyruvate metabolism (239) and starch and sucrose metabolism (218) (Supplementary Table S5).
Table 3.
The top 20 pathways with highest sequence numbers
| Num | Pathway | All genes with pathway annotation (38 200) | Pathway ID |
|---|---|---|---|
| 1 | Metabolic pathways | 4510 (11.81%) | ko01100 |
| 2 | Pathways in cancer | 1790 (4.69%) | ko05200 |
| 3 | Regulation of actin cytoskeleton | 1634 (4.28%) | ko04810 |
| 4 | Focal adhesion | 1518 (3.97%) | ko04510 |
| 5 | MAPK signaling pathway | 1463 (3.83%) | ko04010 |
| 6 | Endocytosis | 1345 (3.52%) | ko04144 |
| 7 | Tight junction | 1256 (3.29%) | ko04530 |
| 8 | Adherens junction | 1073 (2.81%) | ko04520 |
| 9 | Phagosome | 1034 (2.71%) | ko04145 |
| 10 | Dilated cardiomyopathy | 1027 (2.69%) | ko05414 |
| 11 | Vascular smooth muscle contraction | 1014 (2.65%) | ko04270 |
| 12 | Complement and coagulation cascades | 1005 (2.63%) | ko04610 |
| 13 | Hypertrophic cardiomyopathy (HCM) | 957 (2.51%) | ko05410 |
| 14 | Chemokine signaling pathway | 955 (2.5%) | ko04062 |
| 15 | Calcium signaling pathway | 942 (2.47%) | ko04020 |
| 16 | Axon guidance | 939 (2.46%) | ko04360 |
| 17 | Insulin signaling pathway | 912 (2.39%) | ko04910 |
| 18 | Huntington's disease | 907 (2.37%) | ko05016 |
| 19 | Leukocyte transendothelial migration | 869 (2.27%) | ko04670 |
| 20 | Protein processing in endoplasmic reticulum | 864 (2.26%) | ko04141 |
3.4. Positive selection of genes involved in GSH synthesis
Microcystins (MCs) are cyclic non-ribosomal peptides produced by cyanobacteria. They are cyanotoxins and can be very toxic to fishes and other animals, including humans. With the increasing frequency of water bloom outbreaks in many countries, the task of eliminating them has become both more urgent and more difficult. Recently, the silver carp and bighead carp have been used to counteract cyanobacteria in many lakes in China.24,37 Despite the hepatotoxicity of MC, the body weights of silver carp increase very fast in bodies of water that are full of MCs.38 The high tolerance of silver carp to MCs might be due to the high basic GSH level in the liver or an increased GSH synthesis.39
To fully understand the mechanism behind the high tolerance of silver cap to MCs, we evaluated whether the genes involved in glutathione synthesizing were under positive selection in silver carp. From the KEGG databases, 44 genes involved in glutathione synthesis in zebrafish were obtained with the full CDS region (PATHWAY dre00480). After searching against the whole transcriptome sequences, we found that most of the zebrafish CDS had been recovered (Table 4). Sequence pairs were constructed by the zebrafish CDS, and its corresponding best-hit blast scaffolds of silver carp were thereafter tested for whether they had experienced positive selection. For these 44 genes, the average number of codons was found to be 356 (range, 137–966). The F3 × 4 model of codon frequencies was used and models M7 and M8 were used to determine which pairs of sequences were under positive selection. The log-likelihood value under M8 was much higher than its corresponding value under M7, indicating that model M8 is more suitable to the sequence pair compared with model M7. LRT shows that five sequence pairs were found to have P-values <0.05. They are thought to have experienced positive selection between silver carp and zebrafish (Table 5).
Table 4.
Sequences recovered in the glutathione synthesizing pathway
| Gene id | Description | Length | Matched |
|---|---|---|---|
| dre:100002145 | Gamma-glutamyltranspeptidase | 2082 | 0 |
| dre:100006589 | Isocitrate dehydrogenase 1 (NADP+) | 1290 | 1254 |
| dre:100124622 | Glutathione S-transferase | 672 | 514 |
| dre:100330864 | Ribonucleoside-diphosphate reductase subunit M2-like | 1161 | 1057 |
| dre:100333757 | Gamma-glutamyltransferase 5-like | 1521 | 1162 |
| dre:114426 | Ornithine decarboxylase | 1386 | 1370 |
| dre:30733 | Ribonucleotide reductase M2 polypeptide | 1161 | 1057 |
| dre:30740 | Ribonucleotide reductase M1 polypeptide | 2385 | 2385 |
| dre:322533 | Alanyl (membrane) aminopeptidase b | 2898 | 1238 |
| dre:324366 | Glutathione S-transferase M | 660 | 658 |
| dre:324900 | Protein-disulfide reductase (glutathione) | 519 | 515 |
| dre:326857 | Glutamate-cysteine ligase, catalytic subunit | 1896 | 1896 |
| dre:333974 | Glutamate-cysteine ligase, modifier subunit | 822 | 736 |
| dre:352926 | Glutathione peroxidase 1a | 576 | 565 |
| dre:352928 | Glutathione peroxidase 4a | 561 | 561 |
| dre:352929 | Glutathione peroxidase 4b | 576 | 576 |
| dre:386951 | Isocitrate dehydrogenase 2 (NADP+), mitochondrial | 1350 | 1350 |
| dre:394009 | Spermidine synthase | 870 | 749 |
| dre:406278 | Gamma-glutamylcyclotransferase | 663 | 600 |
| dre:406703 | Glutathione S-transferase, alpha-like | 672 | 571 |
| dre:406736 | Glutathione S-transferase | 453 | 425 |
| dre:406762 | Phosphogluconate hydrogenase | 1536 | 1418 |
| dre:431762 | Glutathione S-transferase | 459 | 451 |
| dre:436833 | Glutathione S-transferase kappa 1 | 690 | 680 |
| dre:436894 | Glutathione S-transferase | 723 | 722 |
| dre:449784 | Microsomal glutathione S-transferase 1 | 465 | 244 |
| dre:450084 | Glutathione synthetase | 1428 | 1385 |
| dre:552981 | Glutathione peroxidase 7 | 561 | 0 |
| dre:553169 | Glutathione S-transferase pi 2 | 627 | 625 |
| dre:553575 | Glutathione reductase (NADPH) | 1278 | 1257 |
| dre:555478 | Aminopeptidase N | 2883 | 763 |
| dre:562854 | Leucine aminopeptidase 3 | 1554 | 1320 |
| dre:563972 | Glutathione S-transferase theta 1a | 729 | 729 |
| dre:566746 | Gamma-glutamyltranspeptidase | 1773 | 82 |
| dre:567275 | Glutathione S-transferase | 423 | 423 |
| dre:568744 | Glutathione S-transferase M3 | 660 | 658 |
| dre:569014 | Gamma-glutamyltranspeptidase 1-like | 1725 | 281 |
| dre:570579 | Glucose-6-phosphate dehydrogenase | 1572 | 1572 |
| dre:571365 | Glutathione S-transferase | 660 | 658 |
| dre:723997 | Microsomal glutathione S-transferase 2 | 411 | 0 |
| dre:79381 | Glutathione S-transferase pi | 627 | 626 |
| dre:798788 | Glutathione peroxidase 3 | 669 | 529 |
| dre:799288 | Glutathione S-transferase | 672 | 512 |
| dre:80872 | Spermine synthase | 1083 | 1057 |
Table 5.
Genes determined to be under positive selection
| Gene id | Model | Log likelihood | dN/dS | Estimates of parameters | Sites under selection (P > 0.95) |
|---|---|---|---|---|---|
| dre_322533 | M7(beta) | −4530.079761 | 0.3750 | p = 0.00500 and q = 0.00810 | 164,167,256 |
| M8(beta and ω) | −4519.934204 | 0.7805 | p0 = 0.93767, p = 0.04957, q = 0.14698 and w = 8.71914 | ||
| dre_406703 | M7(beta) | −1299.159488 | 0.1663 | p = 0.13888 and q = 0.68894 | No |
| M8(beta & ω) | −1295.384925 | 16.1965 | p0=0.94248, p=9.04140, q = 99.00000 and w = 280.21751 | ||
| dre_563972 | M7(beta) | −1399.109283 | 0.3750 | p = 0.00542 and q = 0.00912 | 224,226,227,233 |
| M8(beta & ω) | −1390.997418 | 2.5343 | p0 = 0.91793, p = 9.32406, q = 32.38965 and w = 28.38710 | ||
| dre_79381 | M7(beta) | −1105.063900 | 0.1428 | p = 0.03221 and q = 0.18627 | 129,174 |
| M8(beta & ω) | −1100.372249 | 14.0364 | p0 = 0.97094, p = 7.50265, q = 99.00000 and w = 480.64973 | ||
| dre_799288 | M7(beta) | −1334.096144 | 0.1833 | p = 0.12802 and q = 0.56863 | No |
| M8(beta & ω) | −1328.432914 | 13.0557 | p0 = 0.92145, p = 9.04080, q = 99.00000 and w = 165.23531 |
3.5. Identification of SSR or microsatellites
Because SSRs or microsatellite markers are used for many animal breeding applications, the 85 796 sequences were analyzed for identification of SSR markers. We obtained 13 327 SSR markers in 9636 sequences with the MISA.34 In terms of abundance, mononucleotide repeats were found to be most abundant (7693, 57.3%) followed by dinucleotide repeats (3733, 28.0%) and trinucleotide repeats (1538, 11.5%). Other type of repeat units occurred at <2% each. SSR markers were divided into two groups, perfect SSR markers (only one single repeat motif such as ‘AGC’) and compound SSR markers (composed of two or more SSR markers separated by <100 bp). A total of 1206 (9.0%) compound SSR markers were identified. After excluding the mono-nucleotide repeats, the frequency of an SSR motif was calculated. Among the dinucleotide repeat motifs, AC/GT was the most abundant, with 69.2%; trinucleotide repeat motifs were rich in ATC/GAT, with 27.8%, and tetranucleotide repeat motifs were AGAT/ATCT, with 21.1%.
4. Discussion
The transcriptome is the complete repertoire of expressed RNA transcripts in the cell and its characterization is essential to understanding the functional complexity of the genome. Using the next-generation sequencing technology, we were able to sequence and annotate the transcriptome of silver carp. This is most comprehensive study of silver carp transcriptome data to date. The transcriptome sequences obtained by this study are useful to the understanding of the genetic makeup of the silver carp transcriptome, which until now has been very limited. The Illumina sequencing yielded 17.10 million paired-end reads for silver carp. The 85 769 sequences produced here may be useful for further research into silver carp functional genomics. The obtained overall GC content of the silver carp transcriptome was 39.2%, which was lower than the GC content of cDNA library of zebrafish (Ensembl 61).31 However, when we removed the assembled sequences that contained gaps, the GC content rose to 45.5%, which was similar to that of the zebrafish cDNA library (46.2%).
Due to the lack of a complete genome sequence, the quality of transcriptome analysis of non-model species must rely largely on the contigs and scaffolds assembled from the raw reads. After reassembling the transcriptomes of two mosquitoes with known genomes using a de novo assembler, Gibbons et al.40 found that short reads can be used to assemble transcriptomes of non-model organisms. Although the development of the short-read assembler28,41,42 has rendered research facilities capable of dealing with more and more reads, de novo assembly of transcriptomes without known reference genome using short reads is still difficult for transcripts with highly variable coverage.43 So, a higher k-mer length will theoretically generate a more contiguous assembly of highly expressed RNAs while poorly expressed transcripts will be more easily obtained if a lower k-mer length is used.41 Therefore, an approach for de novo assembly of the transcriptome using various k-mer lengths is highly desirable and has been proven useful.27 The final assembly statistics indicate that the multiple k-mer method used in this study outperforms all other single k-mer methods (Table 1). In the single k-mer assembly, the average length and N50 were highest when the k-mer was set to 46, which we found to be best in all single k-mer assemblies. However, the number of contigs >100 bp and total length were twice that of the best single k-mer ABySS assembly. This marked increase was accompanied by a higher N50 and average contig size, indicating a substantial improvement in contiguity.
Multiple k-mer methods assembled the Illumina reads into contigs, but the location information in paired-end reads were not used at all. In this study, we improved upon the methods described by Yann et al.27 Results proved that using SSPACE to scaffold the contigs produced by each k-mer could produce longer sequences. The statistics before and after scaffolding (Tables 1 and 2) indicate that a higher average length of sequence can be obtained by joining the two contigs originating from the two ends of a DNA fragment. The max length and the average length of the sequences after scaffolding were further extended. To assess the quality of assembled transcriptome, we used both computing and experimenting assays to validate the transcripts generated. The RT–PCR results confirmed that our method is reliable for the recovery of both highly and poorly expressed transcripts.
Both gene annotation and KEGG pathway analyses are useful for us to predict potential genes and their functions at a whole-transcriptome level. In the silver carp transcriptome, as discovered by this study, the predominant gene clusters are involved in the structural formation of the cell, cell part and organelle of a cellular component, the binding and catalytic activity of molecular function, metabolic process and cellular processes of biological processes. Similar results were found in Sus scrofa,44 European eel21 and rainbow trout.45 However, in Chickpea transcriptome, sequences were found to be mainly involved in the protein metabolism of biological process, in chloroplasts, in the transferase activity of molecular function. This suggests remarkable difference between animal and plants. KEGG analysis showed that more than 44.5% of transcripts to be enrichment factors involved in 218 known metabolic or signaling pathways, including cell adherence, migration, apoptosis and immune-related processes. The KEGG pathway analysis and gene annotation may be useful for further investigation of gene function in future. Although there are differences between our silver carp transcriptome and available database for zebrafish in GO annotations, concordance in the overall patterns suggests that our library were widely sampled and provided a good representation.
One previous study reported that GSH can conjugate with MC on its sulfhydryl, which is the first step in the detoxification of a cyanobacterial toxin in aquatic organisms.46 The glutathione S-transferase (GST) gene plays important roles both in the biosynthesis of GST and catalysis of the reaction between GSH and MC. M8 assumes 11 site classes: 10 classes for the beta distribution and 1 class for the positively selected site. Therefore, it is suitable to detect positive selection in sequence pairs. Although the value of dN/dS observed in this model might not be precise, the LRT is most likely reliable. Positive selection pressure on GST gene might be the result of the adaptation of silver carp to the eutrophied bodies of water in the middle and lower reaches of the Yangtze River.
Genetic markers are of great importance to the understanding genetic variation and to the identification of genes and quantitative trait locus for traits of interested in molecular breeding applications. Until now, only a small number of genetic markers have been available for silver carp.47,48 One of the main reasons for this is the lack of genome sequence information. Alternatively, transcriptomes have been used for the discovery of genetic markers.44,49,50 Although markers developed from transcriptomes are less polymorphic, they have been found to be very useful in trait mapping51 and comparative genomics studies.52
It has been reported that SSRs comprise 3% of the human genome, with the largest proportion of them being dinucleotide repeats (0.5%).53 In this study, 28% of the 13 327 silver carp SSRs were found to be dinucleotide repeats, followed by trinucleotide repeats (11.5%). The most common dinucleotide repeats were AC and AG, in contrast to those found in the human genome (AC and AT). The same difference was also found in trinucleotide repeats, with ATC and AGG being most common in silver carp and AAT and AAC being most common in human.53
In conclusion, we have determined the transcriptome of silver carp through use of high-throughput Illumina paired-end sequencing. Our study obtained 85 759 scaffolds and demonstrated some important features of silver carp transcriptome, such as gene annotation and KEGG pathway analysis, as shown by cross-transcriptome analysis. In addition, we identified reliable genetic markers for 13 324 SSRs. We also found that five genes identified as under positive selection between silver carp and zebrafish. This study will be helpful for improvement of the understanding of the recent speciation and adaption of Cyprinidae and provides useful resources and markers for future functional genomic research.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This research was supported by the grants from National Basic Research Program of China (973 Program 2010CB126302), National Natural Science Foundation of China (31090254 and U1036603) and Chinese Academy of Sciences (KSCX2-EW-Q-12).
Supplementary Material
References
- 1.Rockman M.V., Kruglyak L. Genetics of global gene expression. Nat. Rev. 2006;7:862–72. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 2.Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 3.Fullwood M.J., Wei C.L., Liu E.T., Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–32. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger M.F., Levin J.Z., Vijayendran K., et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–27. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang G., Guo G., Hu X., et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20:646–54. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu T., Lu G., Fan D., et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–49. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillier L.W., Reinke V., Green P., Hirst M., Marra M.A., Waterston R.H. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–66. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wurtzel O., Sapra R., Chen F., Zhu Y., Simmons B.A., Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–41. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloonan N., Forrest A.R., Kolle G., et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–9. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 10.Lister R., O'Malley R.C., Tonti-Filippini J., et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Q., Shai O., Lee L.J., Frey J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genetics. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 12.Sultan M., Schulz M.H., Richard H., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 13.Johansen S.D., Karlsen B.O., Furmanek T., et al. RNA deep sequencing of the Atlantic cod transcriptome. Comp Biochem Physiol. 2010;6:18–22. doi: 10.1016/j.cbd.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Xiang L.X., He D., Dong W.R., Zhang Y.W., Shao J.Z. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics. 2010;11:472. doi: 10.1186/1471-2164-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson J.S. Fishes of the World. 2006. John Wiley: Hoboken, N.J. [Google Scholar]
- 16.Aparicio S., Chapman J., Stupka E., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–10. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 17.Jaillon O., Aury J.M., Brunet F., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 18.Kasahara M., Naruse K., Sasaki S., et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–9. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 19.Star B., Nederbragt A.J., Jentoft S., et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–10. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser B.A., Weadick C.J., Janowitz I., Rodd F.H., Hughes K.A. Sequencing and characterization of the guppy (Poecilia reticulata) transcriptome. BMC Genomics. 2011;12:202. doi: 10.1186/1471-2164-12-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coppe A., Pujolar J.M., Maes G.E., et al. Sequencing, de novo annotation and analysis of the first Anguilla anguilla transcriptome: EeelBase opens new perspectives for the study of the critically endangered European eel. BMC Genomics. 2010;11:635. doi: 10.1186/1471-2164-11-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Li J., He S. Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol. Phylogenet. Evol. 2007;42:157–70. doi: 10.1016/j.ympev.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Bureau of Fishery; Ministry of Agriculture. China Fishery Statistical Yearbook. Beijing: China Agriculture Press; 2010. [Google Scholar]
- 24.Xie P., Liu J. Practical success of biomanipulation using filter-feeding Fish to control cyanobacteria blooms: a synthesis of decades of research and application in a subtropical hypereutrophic lake. Scientific World J. 2001;1:337–56. doi: 10.1100/tsw.2001.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song W.Q., Libing Z., Jinxin W. Large enclosures experimental study on algal control by silver carp and bighead. Chinese Environ. Sci. 2009;29:1190–5. [Google Scholar]
- 26.Tao W., Zou M., Wang X., Gan X., Mayden R. L., He S. Phylogenomic analysis resolves the formerly intractable adaptive diversification of the endemic clade of east Asian Cyprinidae (Cypriniformes) PLoS One. 2010;5:e13508. doi: 10.1371/journal.pone.0013508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yann Surget-Groba, Montoya-Burgos J.I. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20:1432–40. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J., Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–23. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boetzer M., Henkel C.V., Jansen H.J., Butler D., Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–9. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 30.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 31.Flicek P., Amode M.R., Barrell D., et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–6. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotz S., Garcia-Gomez J.M., Terol J., et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiel T., Michalek W., Varshney R.K., Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor. Appl. Genet. 2003;106:411–22. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 36.Li R., Li Y., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–4. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 37.Ke Z., Xie P., Guo L., Liu Y., Yang H. In situ study on the control of toxic microcystis blooms using phytoplanktivorous fish in the subtropical Lake Taihu of China: a large fish pen experiment. Aquaculture. 2007;265:127–38. [Google Scholar]
- 38.Chen J., Xie P., Zhang D., Ke Z., Yang H. In situ studies on the bioaccumulation of microcystins in the phytoplanktivorous silver carp (Hypophthalmichthys molitrix) stocked in Lake Taihu with dense toxic microcystis blooms. Aquaculture. 2006;261:1026–38. [Google Scholar]
- 39.Li L., Xie P., Li S., Qiu T., Guo L. Sequential ultrastructural and biochemical changes induced in vivo by the hepatotoxic microcystins in liver of the phytoplanktivorous silver carp Hypophthalmichthys molitrix. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007;146:357–67. doi: 10.1016/j.cbpc.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Gibbons J.G., Janson E.M., Hittinger C.T., Johnston M., Abbot P., Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol. Biol. Evol. 2009;26:2731–44. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 41.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren R.L., Sutton G.G., Jones S.J., Holt R.A. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–1. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster S.C. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–8. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 44.Nie Q., Fang M., Jia X., et al. Analysis of muscle and ovary transcriptome of Sus scrofa: assembly, annotation and marker discovery. DNA Res. 2011;18:343–51. doi: 10.1093/dnares/dsr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salem M., Rexroad C.E., 3rd, Wang J., Thorgaard G.H., Yao J. Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics. 2010;11:564. doi: 10.1186/1471-2164-11-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pflugmacher S., Wiegand C., Oberemm A., et al. Identification of an enzymatically formed glutathione conjugate of the cyanobacterial hepatotoxin microcystin-LR: the first step of detoxication. Biochim. Biophys. Acta. 1998;1425:527–33. doi: 10.1016/s0304-4165(98)00107-x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L., Yang G., Guo S., Wei Q., Zou G. Construction of a genetic linkage map for silver carp (Hypophthalmichthys molitrix) Anim. Genet. 2010;41:523–30. doi: 10.1111/j.1365-2052.2010.02031.x. [DOI] [PubMed] [Google Scholar]
- 48.Liao M., Zhang L., Yang G., et al. Development of silver carp (Hypophthalmichthys molitrix) and bighead carp (Aristichthys nobilis) genetic maps using microsatellite and AFLP markers and a pseudo-testcross strategy. Anim. Genet. 2007;38:364–70. doi: 10.1111/j.1365-2052.2007.01617.x. [DOI] [PubMed] [Google Scholar]
- 49.Dubey A., Farmer A., Schlueter J., et al. Defining the transcriptome assembly and its use for genome dynamics and transcriptome profiling studies in pigeonpea (Cajanus cajan L.) DNA Res. 2011;18:153–64. doi: 10.1093/dnares/dsr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dutta S., Kumawat G., Singh B.P., et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh] BMC Plant Biol. 2011;11:17. doi: 10.1186/1471-2229-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W.K., Wang Y.J., Luo G.Z., et al. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor. Appl. Genet. 2004;108:1131–9. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]
- 52.Stein L.D., Bao Z., Blasiar D., et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lander E.S., Linton L.M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.