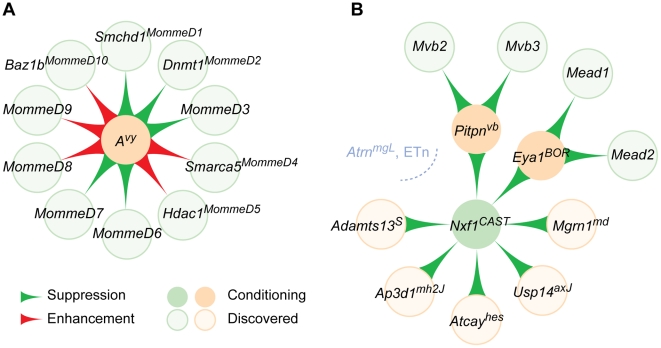
Figure 2. Modifier gene networks have directional edges between nodes.
Mouse modifier gene interactions can be diagrammed as nascent modules of an interaction network. (A) Identification of multiple modifier genes for a conditioning mutation through either mutagenesis or linkage analysis of strain variants can be represented as in “outside-in” network module, where one node (the conditioning mutation, beige circle) is a sink hub, acted on by each experimentally discovered modifier (light green circles). MommeD modifiers of the epigenetically sensitive Avy mutation are illustrated as an example. Direction of effect is indicated by the flared edges (connections) between nodes. (B) Validation of modifier mechanisms across independent mutations result in an “inside-out” module, with the shared modifier (green circle) acting as a source hub on several conditioning or test mutations. An incipient network around Nxf1Mvb1, based on shared genetic mechanism, is illustrated. AtrnmgL and several intronic ETn-induced mutations are not affected by Nxf1Mvb1 variation (gray). Pitpnavb and Eya1BOR have additional known modifiers (light green), adding modules to the incipient network.