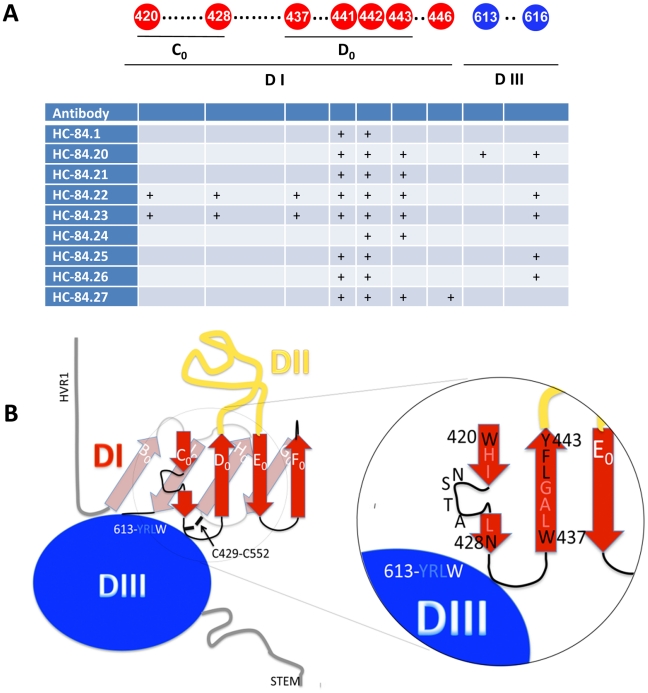
Figure 8. HCV E2 structural analysis.
(A and B) Comparative analysis of the nine HC-84 antibody epitopes and their location on a structure model of the E2 glycoprotein. Contact residues (shown in circles) are located in two discontinuous regions on E2, aa420–446 and aa613–616. Red indicates residues in domain I; black is a cysteine; and blue is residues in domain III. The specific contact residues for the HC-84 antibodies (+) span two domains (I and III) of a structural model of E2, within the central domain I the residues are located on two β-strands, C0, and D0. (B) Mapping of the HC-84 contact residues on the N-terminal side of the 4-stranded C0D0E0F0 β-sheet and more specifically, the C0D0 β-hairpin. To account for the distance between residues 420 and 428, the glycosylation site at 423–425 is shown to be bulging out and with the loop connecting the C0 and D0 strands having a somewhat convoluted 3-dimensional conformation. Residues 428 and 437 are brought close in space in the proposed model.