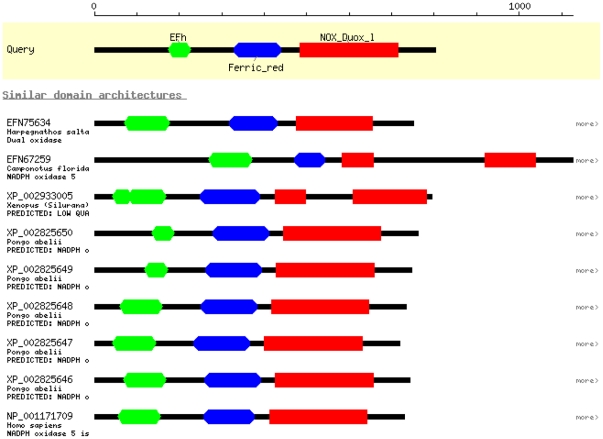
Figure 6. Functional domains and domain architecture based on putative NOX5 sequence.
Based on the alignments, the essential functional residues retrieved from the conserved domain architecture retrieval tool (CDART) showed 10 sequences (coelaomata), 88 sequences (Bilateria), 3 sequences (Deuterostomia), 3 sequences (Magnoliphyta), 65 sequences (cellular organisms), and 120 sequences (embryophyta), in addition to several sequences from other organisms predicted to possess domain architectures similar to that of EF hand (cd00051), Ferric reduct superfamily (pfam01794) and NOX_DuoX_1 (cd06186). Figure illustrating few sequences from different species arranged in a bead_on_string_style showing the presence of predicted EF hand (cd00051), Ferric_reduct super family (pfam01794) and NOX_DuoX_1 (cd06186) domains when rabbit NOX5 was used as query sequence for the domain prediction.