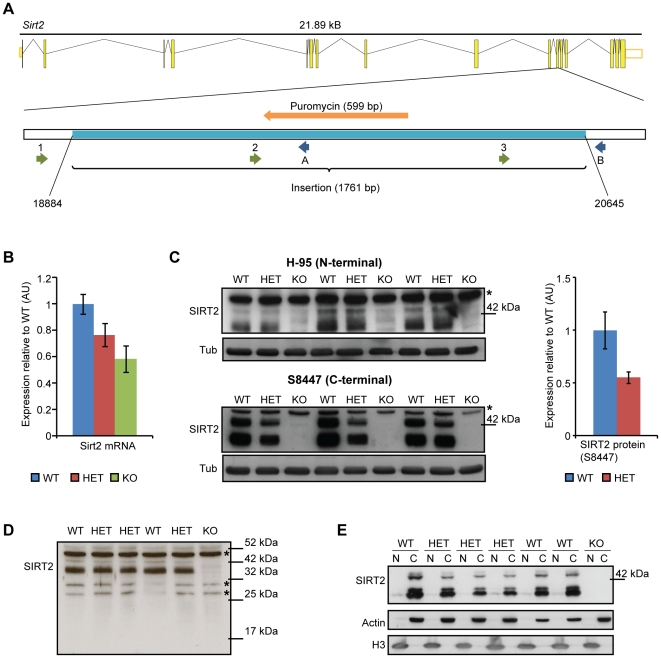
Figure 1. Reduction of Sirt2 mRNA and an absence of the SIRT2 protein in Sirt2 knock-out mice.
(A) Exon-intron structure of the Sirt2 gene in mouse and the location of the insertion (light blue) in exon 11 (after nucleotide 18883) in Sirt2KO mice. The positions of the sequencing forward and reverse primers are shown. 1-Sirt2 forward, 2-Sirt2 forward Seq2, 3-Sirt2 forward Seq3, A-Sirt2 reverse KO, B-Sirt2 reverse WT. (B) Cortical Sirt2 mRNA levels in 4 week old Sirt2KO (KO), Sirt2HET (HET) and wild type (WT) mice. Expression levels were normalised to the housekeeping genes Atp5b and Canx and expressed as fold change of WT levels ±SEM. n = 8/genotype. (C) Western blotting of KO, HET and WT brain lysates with SantaCruz H-95 (upper panel) and Sigma S8447 (lower panel) antibodies. The S8447 probed blot was used to quantify SIRT2 levels (both bands) between HET and WT (right panel). Values were normalised to α-tubulin (Tub) and expressed as fold change of WT ±SEM. * denotes a non-specific band. (D) Western blotting of KO, HET and WT brain lysates with SantaCruz H-95 antibody (long exposure) demonstrating that the Sirt2 disrupting mutation does not result in the production of an N-terminal fragment of SIRT2. *denotes non-specific bands. (E) SIRT2 is localised to the cytoplasm. Purity of fractions was determined by measuring the expression of actin (C-cytoplasm) and H3 (N-nucleus).