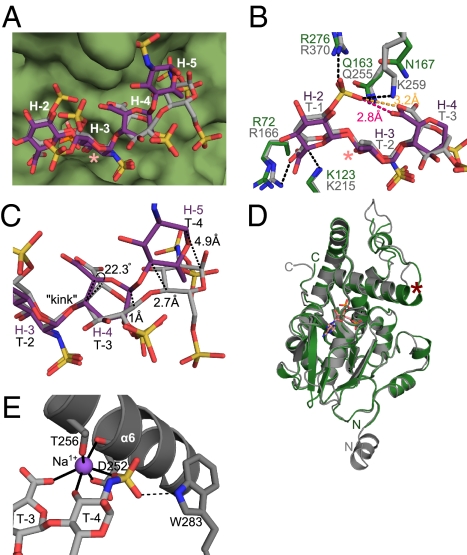
Fig. 4.
Comparison of HS binding to 3-OST-1 and 3-OST-3. (A) Comparison of substrate conformations of the 3-OST-1 bound heptasaccharide (purple) and the 3-OST-3 bound tetrasaccharide (gray, PDB ID code 1T8U) (9). The position of the 3-O-sulfation is labeled with a pink asterisk. The surface of 3-OST-1 is rendered in green. Residues of the heptasaccharide are labeled in white, as in Fig. 3A. (B) Superposition of 3-OST-1 (green) and 3-OST-3 (gray, PDB ID code 1T8U) (9) residues involved in binding their respective oligosaccharide substrates [heptasaccharide (purple) and tetrasaccharide (light gray)]. Putative hydrogen bonding interactions between 3-OST-3 and the tetrasaccharide are shown as dashed lines. The 3-O-sulfation site is labeled with a pink asterisk. (C) Superposition of the heptasaccharide substrate (purple) bound to 3-OST-1 with the tetrasaccharide substrate (gray) bound to 3-OST-3 (PDB ID code 1T8U) (9), highlighting the structural differences between the reducing ends of the chain. (D) Superposition of 3-OST-1 (green) and 3-OST-3 (gray, PDB ID code 1T8U) (9). PAP from the 3-OST-1 structure is drawn in pink. N and C termini are labeled from each respective protein. α-helix 6 helix jutting from the top of the substrate binding cleft is labeled with a red asterisk. (E) Sodium metal ion interactions in the 3-OST-3 substrate binding site. α-helix 6 from 3-OST-3 (9) is shown as a gray ribbon, and the sodium ion as a purple sphere. Interactions coordinating the sodium ion are shown as solid black lines. Potential hydrogen bond between Trp283 and the 6-O-sulfo group on residue T-4 is shown as a dashed line.