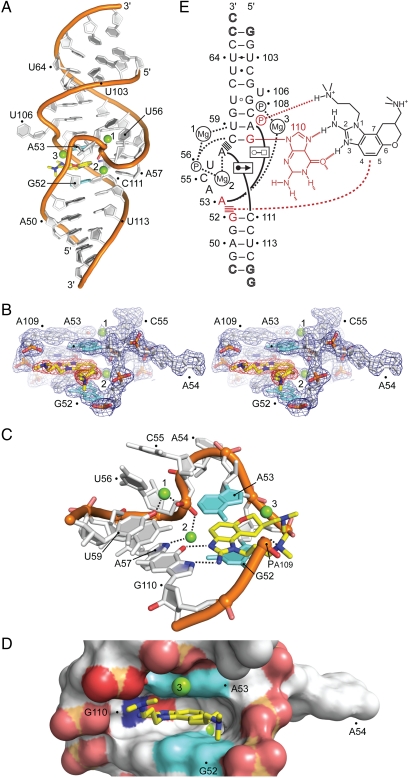
Fig. 2.
Crystal structure of the subdomain IIa RNA inhibitor complex. (A) Overall view of the complex. The benzimidazole ligand (2) is in yellow. Mg2+ ions are shown as green spheres. (B) Stereoview of the 2Fo-Fc electron density contoured at 1σ around the ligand-binding site. (C) Detail view of the ligand-binding site. The bases of G52 and A53, which form the intercalation site for the benzimidazole scaffold, are shown in cyan. The purine of G110 provides the docking edge for the amino-imidazole group. Hydrogen bond interactions are indicated by dashed lines. (D) Surface representation, highlighting the deep ligand-binding pocket. (E) Schematic of the interactions in the ligand-binding site (SI Appendix, Table S3). Hydrogen bonds are shown as dashed lines. Formation of non-Watson–Crick base pairs is indicated with solid lines and symbols according to Leontis et al. (45). Stacked lines (≡) indicate stacking of bases and intercalation of the ligand. Residues interacting with the benzimidazole are highlighted in red.