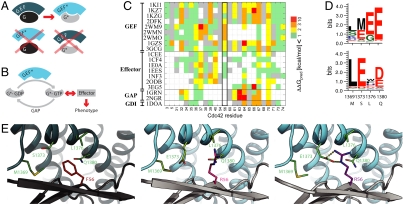
Fig. 1.
Strategy for computational design of an orthogonal signaling interaction. (A) Schematic representation of design requirements for orthogonality: the interface between the GTPase Cdc42 (G) and ITSN (GEF) is modified to generate a pair G*/GEF* with new specificity. (B) Simplified schematic representation of the core GTPase signaling circuit to define the design requirements for a functional G*/GEF* pair that interfaces correctly with other cellular components (GAP and effector proteins that are required for phenotypic output). (C) Computational alanine scanning. Shown are the estimated effects on binding energy of replacing each residue in the Cdc42/ITSN interface (PDB code 1KI1) with alanine in the context of 19 co-complex structures of Cdc42 with partner proteins (white indicates residues not in the interface in the respective structure). Altering position F56 of Cdc42 mainly affects interaction with GEFs. (D) Comparison of fixed backbone (top) and flexible backbone (bottom) computational design predictions for four residues in ITSN (wild-type residues are indicated on the x axis) in the vicinity of position 56 of Cdc42 for a F56R mutation. (E) Model of designed orthoCdc42/orthoITSN interface from fixed (middle) and flexible (right) backbone modeling compared to the wild-type complex (left). Gray: Cdc42; Teal: ITSN; shown in sticks are the five designed interface residues. Small backbone changes modeled by backrub motions (0.53 Å Cα rmsd) allowed the sidechains of R56 and E1373 to adopt conformations that can form hydrogen bonds (dashed lines).