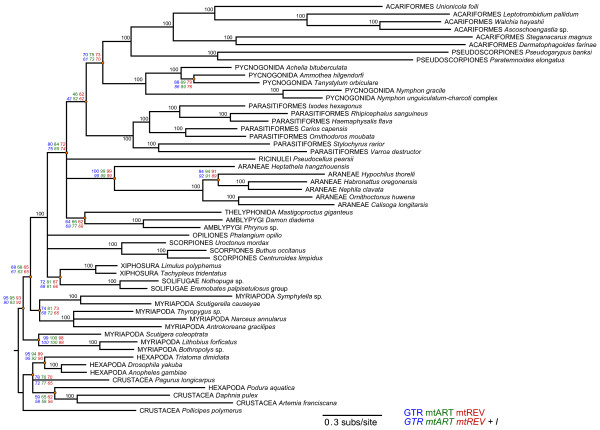
Figure 4.
Consensus tree of 6 different maximum likelihood analyses of amino acid data using different models of evolution. All analyses used the gamma correction model for rate heterogeneity. Bootstrap support values for the 6 analyses are given next to each node. The bootstrap values for the 6 different analyses using 6 models of evolution are given in the following order: Top line = GTR (colored blue); mtART (colored green); mtREV (colored red); Bottom line = GTR + I (colored blue); mtART + I (colored green); mtREV + I (colored red). When all 6 analyses had the same bootstrap value, only one value is given (colored black). Nodes with bootstrap support of less than 60% were manually collapsed. See text for further details of analyses.