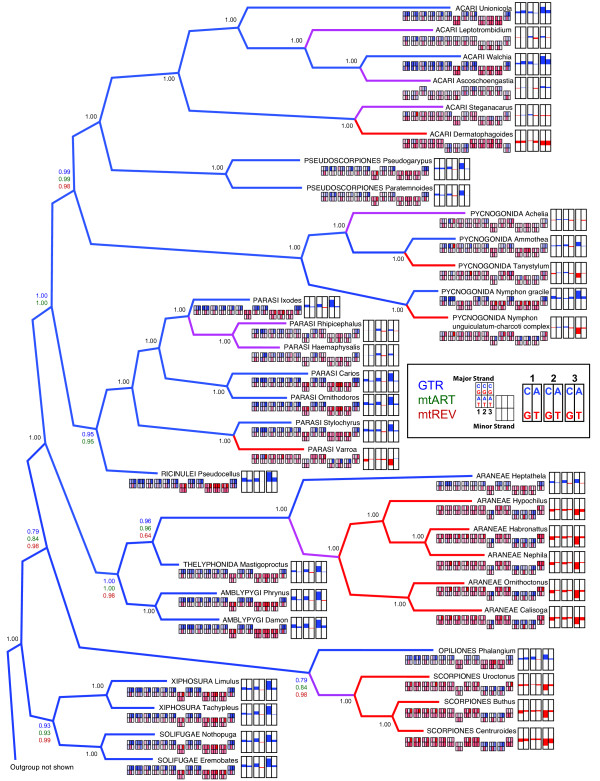
Figure 6.
Phylogenetic tree of chelicerates based on amino acid data, with nucleotide skew shown on the terminal nodes. This Bayesian consensus tree was generated using GTR, mtART, and mtREV models of evolution for amino acids. Site-specific rates across sites were modeled with a Dirichlet process. Posterior probability values are next to the nodes, but when all 3 analyses had the same value, only one value is given (colored black). Outgroup taxa are not depicted. The branches are colored blue if the terminal taxa possess a positive skew towards C or A at 3rd codon positions, red if they possess a negative skew (i.e. G or T skew), or purple if they lack a strong skew (i.e. skew between -0.2 and +0.2). For each terminal taxon, the AT and CG skew of each codon position of the 13 protein-coding genes is shown under the taxon name. The genes are arranged in alphabetical order (ATP6, ATP8, CO1, CO2, CO3, Cytb, ND1, ND2, ND3, ND4, ND4L, ND5 and ND6) to allow gene-by-gene comparisons among otherwise rearranged genomes. Genes located on the minor strand are distinguished by the boxes offset downwards. The skew of each codon position for each of the 13 genes is represented by a color gradient, in which the skew ranges from -1 (red) to 0 (white) to +1 (blue). At the terminal node is a summary of CG and AT skew for each of the codon positions for all 13 genes. Blue depicts cytosine and adenine skew, red depicts guanine and thymine skew. An excess of cytosine is depicted as an upward blue bar, while an excess of guanine is depicted as a downward red bar. An excess of adenine is depicted as an upward blue bar, while an excess of thymine is depicted as a downward red bar.