Table 2. List of the estimated parameter values.
| Parameter | Value | Description |
| β 1 |
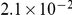
|
Degradation rate of x 1 (based on Western Blot data) |
| β 2 |
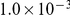
|
Degradation rate of x 2 |
| β 3 |
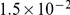
|
Degradation rate of x 3 |
| β 4 |
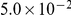
|
Degradation rate of x 4 (based on RT-PCR data) |
| β 5 |

|
Degradation rate of x 5 |
| β 6 |
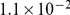
|
Degradation rate of x 6, based on (2) |
| β 7 |
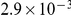
|
Degradation rate of x 7 (based on RT-PCR data) |
| K 1 | 2.1 | Weighted factor |
| K 4 | 11.0 | Weighted factor |
| K 61 | 8.6 | Weighted factor |
| K 66 | 1.3 | Weighted factor |
| K 7 | 1.2 | Weighted factor |
| k 12 |
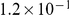
|
Threshold of x 2 to inhibit x 1 |
| k 23 |
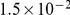
|
Threshold of x 3 to activate x 2 |
| k 41 | 1.7 | Threshold of x 1 to activate x 4 |
| k 61 | 1.4 | Threshold of x 1 to activate x 6 |
| k 66 |
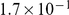
|
Threshold of x 6 to activate x 6 (auto-regulation) |
| k 65 |
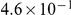
|
Threshold of x 5 to activate x 6 |
| k 76 |
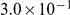
|
Threshold of x 6 to activate x 7 |
| n 12 | 4 | Coefficient of nonlinearity for x 2 to inhibit x 1 |
| n 23 | 4 | Coefficient of nonlinearity for x 3 to activate x 2 |
| n 41 | 4 | Coefficient of nonlinearity for x 1 to activate x 4 |
| n 61 | 4 | Coefficient of nonlinearity for x 1 to activate x 6 |
| n 66 | 4 | Coefficient of nonlinearity for x 6 to activate x 6 |
| n 65 | 4 | Coefficient of nonlinearity for x 5 to activate x 6 |
| n 76 | 4 | Coefficient of nonlinearity for x 6 to activate x 7 |
Parameters such as mRNA degradation rates are estimated based on our data or from existing literatures. Values of other parameters such as activation/inhibition threshold k and relative weights K are estimated to fit in the time course data by genetic algorithm of least square regression. Degradation rates are in the unit of/min. Activation/inhibition thresholds, nonlinearity coefficients and relative weights for multiple inputs are non-dimensionalized.
